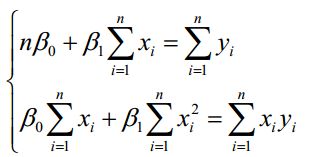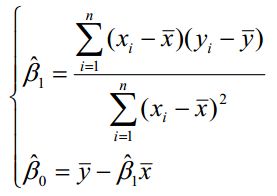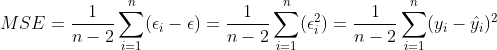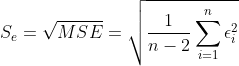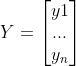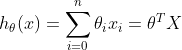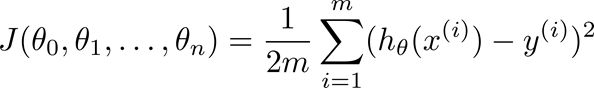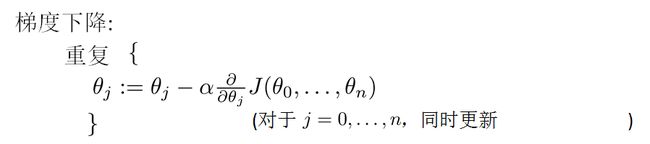SparkML之回归(一)线性回归
----------------------------目录-----------------------------------------------------------------------
线性回归理论
spark源码
Spark实验
-------------------------------------------------------一元线性回归-------------------------------------------------------------------------
模型
反应一个因变量与一个自变量之间的线性关系,一元线性回归模型如下:
![]() (1)
(1)
其中:
![]() 、
、![]() :回归系数
:回归系数
![]() :自变量
:自变量
![]() :因变量
:因变量
![]() :随机误差,一般假设服从
:随机误差,一般假设服从![]()
那么可以得到结论就是:![]() 服从
服从![]()
若我们之前对 (![]() ,
,![]() )进行了 n次观测,那么就可以得到如下,一系列的数据
)进行了 n次观测,那么就可以得到如下,一系列的数据
![]()
![]() 为(1,2,...n)
为(1,2,...n)
那么把這些数值,带入(1)公式,那么就有 n个包含![]() 、
、![]() 方程,大家知道当要确定n个参数的时候,满秩的情况下,只要n个方程就就可以确定了,那么如何根据历史的观测数据来选择,来选择最佳的
方程,大家知道当要确定n个参数的时候,满秩的情况下,只要n个方程就就可以确定了,那么如何根据历史的观测数据来选择,来选择最佳的![]() 、
、![]() ,只要把
,只要把![]() 、
、![]() 确定了,那么我们随便输入一个
确定了,那么我们随便输入一个![]() ,就可以得到一个
,就可以得到一个![]() ,那么选择一个"未来"的
,那么选择一个"未来"的![]() ,就可以计算一个"未来"的
,就可以计算一个"未来"的![]() ,那么就达到了预测效果
,那么就达到了预测效果
普通最小二乘法
那么什么才是最佳的 ![]() 、
、![]() ,最小二乘法的思想就是把决定后的方程,代入参数使得方差最小,就是最佳的。我们把全部的方差记为:
,最小二乘法的思想就是把决定后的方程,代入参数使得方差最小,就是最佳的。我们把全部的方差记为:
![]()
那么现在就是计算![]() 关于参数
关于参数![]() 、
、![]() 的极小值,当
的极小值,当![]() 关于参数
关于参数![]() 、
、![]() 的偏导为0的时候,那么
的偏导为0的时候,那么![]() 取到极值
取到极值
对其进行整理,得到如下:
那么可以直接计算出:
当自变量x多的时候,就很难直接计算![]() 、
、![]() 、....、
、....、![]() ,那么就必须用克拉姆法则(Cramer's Rule)计算,
,那么就必须用克拉姆法则(Cramer's Rule)计算,
其中![]() ,
,![]() 、、、
、、、![]() 是
是![]() 、
、![]() 、....、
、....、![]() 的最小二乘估计。
的最小二乘估计。
拟合效果分析
1、残差的样本方差
残差:![]() (i = 1,2,...n)
(i = 1,2,...n)
其中n-2是自由度,因为有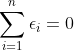 和
和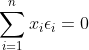 约束,所以自由度减2(残差之间相互独立,残差和自变量x相互独立),如果我们的拟合方程:
约束,所以自由度减2(残差之间相互独立,残差和自变量x相互独立),如果我们的拟合方程:![]() 解释因变量
解释因变量![]() 越强,那么MSE是越小。你会发现:
越强,那么MSE是越小。你会发现:
这个MSE就是总体回归模型中方差的无偏估计量。
2、判定系数(R)
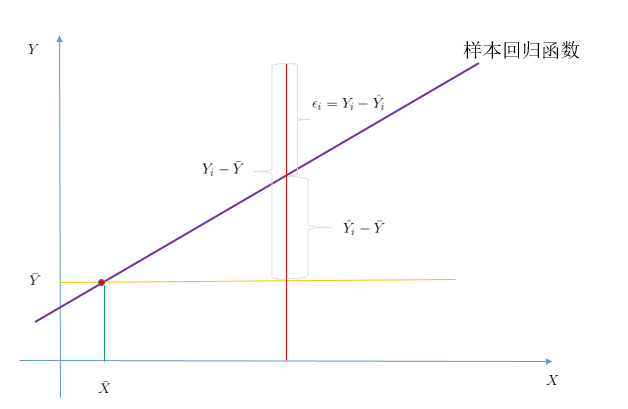
我们从新考虑我们的样本回归函数:
![]()
因为我们的解释变量的平均值![]() ,一定会经过我们的样本回归函数,下面证明:
,一定会经过我们的样本回归函数,下面证明:
![]()
两边进行平方之后再加总,然后除以样本容量n:
下面结合图像进行说明:
结合图像,我们可以得到下面方程:![]()
两边平方之后,进行加总,得到:
![]()
![]() :样本观测值和其平均值的离差平方和,自由度为n-1
:样本观测值和其平均值的离差平方和,自由度为n-1
![]() :拟合直线
:拟合直线![]() 可解释部分的平方和,自由度为1
可解释部分的平方和,自由度为1
![]() :样本的观测值和估计值之差的平方,既残差平方和,自由度为n-2
:样本的观测值和估计值之差的平方,既残差平方和,自由度为n-2
缩写全拼(采用国外教材的缩写方式):
Total sum of squares(SST):总离差平方和
Residual sum of squares (SSR):残差平方和
explained sum of squares(SSE):回归平方和(国人根据实际意义自己命名的?)
所以我们有:![]()
那么对于我们真正解释了的部分和总体的比值(用![]() 表示):
表示):
当![]() 时,也就是SSR = SSE,那么就是说原始数据完全可以拟合值来解释,此时SSR = 0,那么拟合非常完美
时,也就是SSR = SSE,那么就是说原始数据完全可以拟合值来解释,此时SSR = 0,那么拟合非常完美
一般![]() 。
。
SSR很好计算,就是样本的实际观察值与估计值差的平方,所以用SSR去计算R
显著性检验
当你拟合好参数的时候,你要去评定一个這样的一个模型对于我们想要解释的问题是否显著(只有R是不够的),
如果不显著那么就需要换其他模型方法了。对于其中检验的方法有F检验和T检验,本文重点是SparkMlib下的线性回归,本节只是一个铺垫,所以具体如何检验,就不赘述了。
-------------------------------------------------------多元线性回归----------------------------------------------------------------------------
模型
反应多个因变量与一个自变量之间的线性关系,多元线性回归模型如下:
其中:![]() ,
,![]() 都是与
都是与![]() 无关的未知参数,
无关的未知参数,![]() 是回归系数。
是回归系数。
现在得到n个样本数据(![]() ),
),![]() =1,....,n,其中
=1,....,n,其中![]() ,那么(2)得到:
,那么(2)得到:
我们可以把(3)写成如下模式:
其中:
求解![]() 过程和一元线性回归一样,可以得到:
过程和一元线性回归一样,可以得到:
![]()
判定系数(R)还是按照一元回归那样求解,当R大于0.8才认为线性关系明显
===================================最小二乘法的缺陷============================
1、只有当X满秩的时候,才可以用最小二乘法。因为在求解的时候的条件:X是满秩的,也就是在决定多个因变量
![]() 必须是相互独立的,当如果
必须是相互独立的,当如果![]() 和
和![]() 有关联,
有关联,![]() 可以用
可以用![]() 表示,那么X就不是满秩的
表示,那么X就不是满秩的
此时用最小二乘法就是错误的,因为X是不可逆的
2、最小二乘的复杂度高,在处理大规模数据的时候,耗时长。
--------------------------------------------------------------------梯度下降法-------------------------------------------------------------------
由于最小二乘法在求解![]() 时,存在局限,所以在计算机领域一般采用梯度下降法,来近似求解
时,存在局限,所以在计算机领域一般采用梯度下降法,来近似求解![]()
为了与文献2的符号一致,所以放弃前面用过的符号,采用文献2中的符号。现在直接从多元线性回归开始
线性方程:
我们让![]() ,那么方程变为:
,那么方程变为:
若我们之前对 (![]() ,
,![]() )进行了 m次观测,那么就可以得到如下,一系列的数据
)进行了 m次观测,那么就可以得到如下,一系列的数据
![]()
![]() 为(1,2,...m),按照前面的思路,我们来计算“相差”多少,既所说的cost function:
为(1,2,...m),按照前面的思路,我们来计算“相差”多少,既所说的cost function:
(小插曲:不知道为什么有很多人把上面的m给省略了,在andrew NG课程中和Spark源码理解中都有这个m
其实加上m更能体现问题)
也就说让![]() 最小。如果用之前的最小二乘法,那么就是,让
最小。如果用之前的最小二乘法,那么就是,让![]() 对
对![]() 求偏导,让等式都等于0,建立方程,联合求解
求偏导,让等式都等于0,建立方程,联合求解![]() :
:
![]()
我们知道最小二乘法的弊端,所以采用梯度下降法来求解最优的![]() :
:
其中![]() 是学习效率,而且迭代的初始值
是学习效率,而且迭代的初始值![]() 设置为n+1列的零向量,然后一直迭代,直到收敛为止。
设置为n+1列的零向量,然后一直迭代,直到收敛为止。
当样本很大的时候,如果迭代次数很大,那么我们会选择一部分样本进行对![]() 的更新计算。
的更新计算。
更多细节,请看:http://blog.csdn.net/legotime/article/details/51277141
-------------------------------------------------------------------------------------------------------------------------------------------------
Spark源码
package org.apache.spark.mllib.regression包含了两个部分:LinearRegressionModel和LinearRegressionWithSGD
1、回归的模型(class和object),class 的参数是继承GeneralizedLinearModel广义回归模型,之后形成一个完整的
线性回归模型,object上面的方法用于导出已经保存的模型进行回归
2、LinearRegressionWithSGD:随机梯度下降法,cost function:f(weights) = 1/n ||A weights-y||^2也就是前面
记住这个还是加上m更能体现问题,(除以m表示均方误差)
LinearRegressionWithSGD是继承GeneralizedLinearAlgorithm[LinearRegressionModel]广义回归类
1、回归模型源码如下
/** * Regression model trained using LinearRegression. * * @param weights Weights computed for every feature.(每个特征的权重向量) * @param intercept Intercept computed for this model.(此模型的偏置或残差) * */ @Since("0.8.0") class LinearRegressionModel @Since("1.1.0") ( @Since("1.0.0") override val weights: Vector, @Since("0.8.0") override val intercept: Double) extends GeneralizedLinearModel(weights, intercept) with RegressionModel with Serializable with Saveable with PMMLExportable { //进行预测:Y = W*X+intercept override protected def predictPoint( dataMatrix: Vector, weightMatrix: Vector, intercept: Double): Double = { weightMatrix.toBreeze.dot(dataMatrix.toBreeze) + intercept } //模型保存包含:保存的位置,名字,权重和偏置 @Since("1.3.0") override def save(sc: SparkContext, path: String): Unit = { GLMRegressionModel.SaveLoadV1_0.save(sc, path, this.getClass.getName, weights, intercept) } override protected def formatVersion: String = "1.0" } //加载上面保存和的模型,用load(sc,存储路径) @Since("1.3.0") object LinearRegressionModel extends Loader[LinearRegressionModel] { @Since("1.3.0") override def load(sc: SparkContext, path: String): LinearRegressionModel = { val (loadedClassName, version, metadata) = Loader.loadMetadata(sc, path) // Hard-code class name string in case it changes in the future val classNameV1_0 = "org.apache.spark.mllib.regression.LinearRegressionModel" (loadedClassName, version) match { case (className, "1.0") if className == classNameV1_0 => val numFeatures = RegressionModel.getNumFeatures(metadata) val data = GLMRegressionModel.SaveLoadV1_0.loadData(sc, path, classNameV1_0, numFeatures) new LinearRegressionModel(data.weights, data.intercept) case _ => throw new Exception( s"LinearRegressionModel.load did not recognize model with (className, format version):" + s"($loadedClassName, $version). Supported:\n" + s" ($classNameV1_0, 1.0)") } } }
2、LinearRegressionWithSGD类,该类是基于无正规化的随机梯度下降,而且是继承GeneralizedLinearAlgorithm[LinearRegressionModel]广义回归类
/** * Train a linear regression model with no regularization using Stochastic Gradient Descent. * This solves the least squares regression formulation * f(weights) = 1/n ||A weights-y||^2^ * (which is the mean squared error). * Here the data matrix has n rows, and the input RDD holds the set of rows of A, each with * its corresponding right hand side label y. * See also the documentation for the precise formulation. */ @Since("0.8.0") class LinearRegressionWithSGD private[mllib] ( private var stepSize: Double,//步长 private var numIterations: Int,//迭代次数 private var miniBatchFraction: Double)//参与迭代样本的比列 extends GeneralizedLinearAlgorithm[LinearRegressionModel] with Serializable { private val gradient = new LeastSquaresGradient() //阅读:3 private val updater = new SimpleUpdater() //阅读:4 @Since("0.8.0") override val optimizer = new GradientDescent(gradient, updater) //阅读:5 .setStepSize(stepSize) .setNumIterations(numIterations) .setMiniBatchFraction(miniBatchFraction) /** * Construct a LinearRegression object with default parameters: {stepSize: 1.0, * numIterations: 100, miniBatchFraction: 1.0}. */ @Since("0.8.0") def this() = this(1.0, 100, 1.0) override protected[mllib] def createModel(weights: Vector, intercept: Double) = { new LinearRegressionModel(weights, intercept) } } /** * Top-level methods for calling LinearRegression. * */ @Since("0.8.0") object LinearRegressionWithSGD { /** * Train a Linear Regression model given an RDD of (label, features) pairs. We run a fixed number * of iterations of gradient descent using the specified step size. Each iteration uses * `miniBatchFraction` fraction of the data to calculate a stochastic gradient. The weights used * in gradient descent are initialized using the initial weights provided. * * @param input RDD of (label, array of features) pairs. Each pair describes a row of the data * matrix A as well as the corresponding right hand side label y * @param numIterations Number of iterations of gradient descent to run. * @param stepSize Step size to be used for each iteration of gradient descent. * @param miniBatchFraction Fraction of data to be used per iteration. * @param initialWeights Initial set of weights to be used. Array should be equal in size to * the number of features in the data. * */ @Since("1.0.0") def train( input: RDD[LabeledPoint], numIterations: Int, stepSize: Double, miniBatchFraction: Double, initialWeights: Vector): LinearRegressionModel = { new LinearRegressionWithSGD(stepSize, numIterations, miniBatchFraction) .run(input, initialWeights) } /** * Train a LinearRegression model given an RDD of (label, features) pairs. We run a fixed number * of iterations of gradient descent using the specified step size. Each iteration uses * `miniBatchFraction` fraction of the data to calculate a stochastic gradient. * * @param input RDD of (label, array of features) pairs. Each pair describes a row of the data * matrix A as well as the corresponding right hand side label y * @param numIterations Number of iterations of gradient descent to run. * @param stepSize Step size to be used for each iteration of gradient descent. * @param miniBatchFraction Fraction of data to be used per iteration. * */ @Since("0.8.0") def train( input: RDD[LabeledPoint], numIterations: Int, stepSize: Double, miniBatchFraction: Double): LinearRegressionModel = { new LinearRegressionWithSGD(stepSize, numIterations, miniBatchFraction).run(input) } /** * Train a LinearRegression model given an RDD of (label, features) pairs. We run a fixed number * of iterations of gradient descent using the specified step size. We use the entire data set to * compute the true gradient in each iteration. * * @param input RDD of (label, array of features) pairs. Each pair describes a row of the data * matrix A as well as the corresponding right hand side label y * @param stepSize Step size to be used for each iteration of Gradient Descent. * @param numIterations Number of iterations of gradient descent to run. * @return a LinearRegressionModel which has the weights and offset from training. * */ @Since("0.8.0") def train( input: RDD[LabeledPoint], numIterations: Int, stepSize: Double): LinearRegressionModel = { train(input, numIterations, stepSize, 1.0) } /** * Train a LinearRegression model given an RDD of (label, features) pairs. We run a fixed number * of iterations of gradient descent using a step size of 1.0. We use the entire data set to * compute the true gradient in each iteration. * * @param input RDD of (label, array of features) pairs. Each pair describes a row of the data * matrix A as well as the corresponding right hand side label y * @param numIterations Number of iterations of gradient descent to run. * @return a LinearRegressionModel which has the weights and offset from training. * */ @Since("0.8.0") def train( input: RDD[LabeledPoint], numIterations: Int): LinearRegressionModel = { train(input, numIterations, 1.0, 1.0) } }
3、最小平方梯度,首先联系我们的代价(损失)函数,如下:
损失函数源码标记为:L = 1/2n ||A weights-y||^2
每个样本的梯度值:![]()
第一个compute返回的是 ![]() ,第二个compute返回的是
,第二个compute返回的是![]()
class LeastSquaresGradient extends Gradient { override def compute(data: Vector, label: Double, weights: Vector): (Vector, Double) = { val diff = dot(data, weights) - label val loss = diff * diff / 2.0//误差 val gradient = data.copy scal(diff, gradient)////梯度值x*(y-h(x)) (gradient, loss) } override def compute( data: Vector, label: Double, weights: Vector, cumGradient: Vector): Double = { val diff = dot(data, weights) - label//h(x)-y axpy(diff, data, cumGradient)//y = x*(h(x)-y)+cumGradient /**axpy用法: * Computes y += x * a, possibly doing less work than actually doing that operation * def axpy[A, X, Y](a: A, x: X, y: Y)(implicit axpy: CanAxpy[A, X, Y]) { axpy(a,x,y) } */ diff * diff / 2.0 } }
4、权重更新(SimpleUpdater),更新公式如下:
返回的时候偏置项设置为0了
class SimpleUpdater extends Updater { override def compute( weightsOld: Vector,//上一次计算后的权重向量 gradient: Vector,//本次迭代的权重向量 stepSize: Double,//步长 iter: Int,//当前迭代次数 regParam: Double): (Vector, Double) = { val thisIterStepSize = stepSize / math.sqrt(iter)//学习速率 a val brzWeights: BV[Double] = weightsOld.toBreeze.toDenseVector brzAxpy(-thisIterStepSize, gradient.toBreeze, brzWeights) //brzWeights + = gradient.toBreeze-thisIterStepSize (Vectors.fromBreeze(brzWeights), 0) } }
5权重优化
权重优化采用的是随机梯度降,但是默认的是miniBatchFraction= 1.0。
/** * Class used to solve an optimization problem using Gradient Descent. * @param gradient Gradient function to be used. * @param updater Updater to be used to update weights after every iteration. */ class GradientDescent private[spark] (private var gradient: Gradient, private var updater: Updater) extends Optimizer with Logging { private var stepSize: Double = 1.0 private var numIterations: Int = 100 private var regParam: Double = 0.0 private var miniBatchFraction: Double = 1.0 private var convergenceTol: Double = 0.001//收敛公差 /** * Set the initial step size of SGD for the first step. Default 1.0. * In subsequent steps, the step size will decrease with stepSize/sqrt(t) */ def setStepSize(step: Double): this.type = { this.stepSize = step this } /** * :: Experimental :: * Set fraction of data to be used for each SGD iteration. * Default 1.0 (corresponding to deterministic/classical gradient descent) */ @Experimental def setMiniBatchFraction(fraction: Double): this.type = { this.miniBatchFraction = fraction this } /** * Set the number of iterations for SGD. Default 100. */ def setNumIterations(iters: Int): this.type = { this.numIterations = iters this } /** * Set the regularization parameter. Default 0.0. */ def setRegParam(regParam: Double): this.type = { this.regParam = regParam this } /** * Set the convergence tolerance. Default 0.001 * convergenceTol is a condition which decides iteration termination. * The end of iteration is decided based on below logic. * * - If the norm of the new solution vector is >1, the diff of solution vectors * is compared to relative tolerance which means normalizing by the norm of * the new solution vector. * - If the norm of the new solution vector is <=1, the diff of solution vectors * is compared to absolute tolerance which is not normalizing. * * Must be between 0.0 and 1.0 inclusively. */ def setConvergenceTol(tolerance: Double): this.type = { require(0.0 <= tolerance && tolerance <= 1.0) this.convergenceTol = tolerance this } /** * Set the gradient function (of the loss function of one single data example) * to be used for SGD. */ def setGradient(gradient: Gradient): this.type = { this.gradient = gradient this } /** * Set the updater function to actually perform a gradient step in a given direction. * The updater is responsible to perform the update from the regularization term as well, * and therefore determines what kind or regularization is used, if any. */ def setUpdater(updater: Updater): this.type = { this.updater = updater this } /** * :: DeveloperApi :: * Runs gradient descent on the given training data. * @param data training data * @param initialWeights initial weights * @return solution vector */ @DeveloperApi def optimize(data: RDD[(Double, Vector)], initialWeights: Vector): Vector = { val (weights, _) = GradientDescent.runMiniBatchSGD( data, gradient, updater, stepSize, numIterations, regParam, miniBatchFraction, initialWeights, convergenceTol) weights } } /** * :: DeveloperApi :: * Top-level method to run gradient descent. */ @DeveloperApi object GradientDescent extends Logging { /** * Run stochastic gradient descent (SGD) in parallel using mini batches. * In each iteration, we sample a subset (fraction miniBatchFraction) of the total data * in order to compute a gradient estimate. * Sampling, and averaging the subgradients over this subset is performed using one standard * spark map-reduce in each iteration. * * @param data Input data for SGD. RDD of the set of data examples, each of * the form (label, [feature values]). * @param gradient Gradient object (used to compute the gradient of the loss function of * one single data example) * @param updater Updater function to actually perform a gradient step in a given direction. * @param stepSize initial step size for the first step * @param numIterations number of iterations that SGD should be run. * @param regParam regularization parameter * @param miniBatchFraction fraction of the input data set that should be used for * one iteration of SGD. Default value 1.0. * @param convergenceTol Minibatch iteration will end before numIterations if the relative * difference between the current weight and the previous weight is less * than this value. In measuring convergence, L2 norm is calculated. * Default value 0.001. Must be between 0.0 and 1.0 inclusively. * @return A tuple containing two elements. The first element is a column matrix containing * weights for every feature, and the second element is an array containing the * stochastic loss computed for every iteration. */ def runMiniBatchSGD( data: RDD[(Double, Vector)], gradient: Gradient, updater: Updater, stepSize: Double, numIterations: Int, regParam: Double, miniBatchFraction: Double, initialWeights: Vector, convergenceTol: Double): (Vector, Array[Double]) = { // convergenceTol should be set with non minibatch settings if (miniBatchFraction < 1.0 && convergenceTol > 0.0) { logWarning("Testing against a convergenceTol when using miniBatchFraction " + "< 1.0 can be unstable because of the stochasticity in sampling.") } //把历史的权重放在一个数组中 val stochasticLossHistory = new ArrayBuffer[Double](numIterations) // Record previous weight and current one to calculate solution vector difference //初始化权重 var previousWeights: Option[Vector] = None var currentWeights: Option[Vector] = None //训练的样本数 val numExamples = data.count() // if no data, return initial weights to avoid NaNs if (numExamples == 0) { logWarning("GradientDescent.runMiniBatchSGD returning initial weights, no data found") return (initialWeights, stochasticLossHistory.toArray) } if (numExamples * miniBatchFraction < 1) { logWarning("The miniBatchFraction is too small") } // Initialize weights as a column vector var weights = Vectors.dense(initialWeights.toArray) val n = weights.size /** * For the first iteration, the regVal will be initialized as sum of weight squares * if it's L2 updater; for L1 updater, the same logic is followed. */ var regVal = updater.compute( weights, Vectors.zeros(weights.size), 0, 1, regParam)._2 var converged = false // indicates whether converged based on convergenceTol判断是否收敛 var i = 1 while (!converged && i <= numIterations) { //广播weights val bcWeights = data.context.broadcast(weights) // Sample a subset (fraction miniBatchFraction) of the total data // compute and sum up the subgradients on this subset (this is one map-reduce) val (gradientSum, lossSum, miniBatchSize) = data.sample(false, miniBatchFraction, 42 + i) .treeAggregate((BDV.zeros[Double](n), 0.0, 0L))( seqOp = (c, v) => { // c: (grad, loss, count), v: (label, features) val l = gradient.compute(v._2, v._1, bcWeights.value, Vectors.fromBreeze(c._1)) (c._1, c._2 + l, c._3 + 1) }, combOp = (c1, c2) => { // c: (grad, loss, count) (c1._1 += c2._1, c1._2 + c2._2, c1._3 + c2._3) }) if (miniBatchSize > 0) { /** * lossSum is computed using the weights from the previous iteration * and regVal is the regularization value computed in the previous iteration as well. */ //保存误差,更新权重 stochasticLossHistory.append(lossSum / miniBatchSize + regVal) val update = updater.compute( weights, Vectors.fromBreeze(gradientSum / miniBatchSize.toDouble), stepSize, i, regParam) weights = update._1 regVal = update._2 previousWeights = currentWeights currentWeights = Some(weights) if (previousWeights != None && currentWeights != None) { converged = isConverged(previousWeights.get, currentWeights.get, convergenceTol) } } else { logWarning(s"Iteration ($i/$numIterations). The size of sampled batch is zero") } i += 1 } logInfo("GradientDescent.runMiniBatchSGD finished. Last 10 stochastic losses %s".format( stochasticLossHistory.takeRight(10).mkString(", "))) //返回权重和历史误差数组 (weights, stochasticLossHistory.toArray) }
SparkML实验:
package Regression import org.apache.spark.mllib.linalg.Vectors import org.apache.spark.mllib.regression.{LabeledPoint, LinearRegressionModel, LinearRegressionWithSGD} import org.apache.spark.{SparkConf, SparkContext} object RegressionWithSGD { def main(args: Array[String]) { val conf = new SparkConf().setAppName("LinearRegressionWithSGDExample").setMaster("local") val sc = new SparkContext(conf) // Load and parse the data val data = sc.textFile("E:\\SparkCore2\\data\\mllib\\ridge-data\\lpsa.data") val parsedData = data.map { line => val parts = line.split(',') LabeledPoint(parts(0).toDouble, Vectors.dense(parts(1).split(' ').map(_.toDouble))) } /**parsedData形式: * (-0.4307829,[-1.63735562648104,-2.00621178480549,-1.86242597251066,-1.02470580167082,-0.522940888712441, * -0.863171185425945,-1.04215728919298,-0.864466507337306]) */ // Building the model val numIterations = 100//迭代次数 val stepSize = 0.00000001//步长 val model = LinearRegressionWithSGD.train(parsedData, numIterations, stepSize)//训练模型 // Evaluate model on training examples and compute training error val valuesAndPreds = parsedData.map { point => val prediction = model.predict(point.features) (point.label, prediction) } val numCount = valuesAndPreds.count() println("The sample count"+numCount) val MSE = valuesAndPreds.map{ case(v, p) => math.pow((v - p), 2) }.mean()//残差的样本方差 println("training Mean Squared Error = " + MSE) println("模型的权重"+model.weights) println("模型的残差"+model.intercept) // Save and load model model.save(sc, "E:\\SparkCore2\\data\\mllib\\ridge-data\\scalaLinearRegressionWithSGDModel") val sameModel = LinearRegressionModel.load(sc, "E:\\SparkCore2\\data\\mllib\\ridge-data\\scalaLinearRegressionWithSGDModel") sc.stop() /** * The sample count:67 * training Mean Squared Error = 7.4510328101026 *模型的权重[1.440209460949548E-8,1.0686674736254139E-8,9.608973495307957E-9,4.553409983798095E-9,1.2221496560765207E-8,8.910773406981891E-9,5.5962085583952E-9,1.2255699128757168E-8] *模型的残差0.0 */ } }
参考文献:
1andrew NG线性回归课件:链接:http://pan.baidu.com/s/1bTgHgq 密码:7mbt