神经网络&DNN算法原理及代码实现(更新中)
本文整理了神经网络&DNN算法原理及代码实现,主要参考了吴恩达老师的深度学习课程以及课程作业,适合中级学习人员做参考资料查阅
1.Normalizing inputs(归一化/标准化输入)
the weight matrices ( W [ 1 ] , W [ 2 ] , W [ 3 ] , . . . , W [ L − 1 ] , W [ L ] ) (W^{[1]}, W^{[2]}, W^{[3]}, ..., W^{[L-1]}, W^{[L]}) (W[1],W[2],W[3],...,W[L−1],W[L]),the bias vectors ( b [ 1 ] , b [ 2 ] , b [ 3 ] , . . . , b [ L − 1 ] , b [ L ] ) (b^{[1]}, b^{[2]}, b^{[3]}, ..., b^{[L-1]}, b^{[L]}) (b[1],b[2],b[3],...,b[L−1],b[L])
- min-max标准化
x ′ = x − min ( x ) max ( x ) − min ( x ) x^{\prime}=\frac{x-\min (x)}{\max (x)-\min (x)} x′=max(x)−min(x)x−min(x)
def min_max_normalization(X):
X = float(X - np.min(X))/(np.max(X)- np.min(X))
return X
- Z-score标准化
x ∗ = x − μ σ x^{*}=\frac{x-\mu}{\sigma} x∗=σx−μ
def min_max_normalization(X):
X = float(X - np.min(X))/(np.max(X)- np.min(X))
return X
2. Initialize_parameters(初始化参数W, b)
- Zero initialization
W, b = np.zeros((…, …))
def initialize_parameters_zeros(layers_dims):
"""
Arguments:
layer_dims -- python array (list) containing the size of each layer.
Returns:
parameters -- python dictionary containing your parameters "W1", "b1", ..., "WL", "bL":
W1 -- weight matrix of shape (layers_dims[1], layers_dims[0])
b1 -- bias vector of shape (layers_dims[1], 1)
...
WL -- weight matrix of shape (layers_dims[L], layers_dims[L-1])
bL -- bias vector of shape (layers_dims[L], 1)
"""
parameters = {}
L = len(layers_dims) # number of layers in the network
for l in range(1, L):
parameters['W' + str(l)] = np.zeros((layers_dims[l],layers_dims[l-1]))
parameters['b' + str(l)] = np.zeros((layers_dims[l],1))
return parameters
- Random initialization
W,b = np.random.randn((…, …))*10
def initialize_parameters_random(layers_dims):
"""
Arguments:
layer_dims -- python array (list) containing the size of each layer.
Returns:
parameters -- python dictionary containing your parameters "W1", "b1", ..., "WL", "bL":
W1 -- weight matrix of shape (layers_dims[1], layers_dims[0])
b1 -- bias vector of shape (layers_dims[1], 1)
...
WL -- weight matrix of shape (layers_dims[L], layers_dims[L-1])
bL -- bias vector of shape (layers_dims[L], 1)
"""
parameters = {}
L = len(layers_dims) # integer representing the number of layers
for l in range(1, L):
parameters['W' + str(l)] = np.random.randn(layers_dims[l],layers_dims[l-1])*10
parameters['b' + str(l)] = np.zeros((layers_dims[l],1))
return parameters
- He initialization
w [ l ] = n p . w^{[l]}=n p . w[l]=np. random. randn (shape) * np. sqrt ( 2 n [ l − 1 ] ) \left(\frac{2}{n^{[l-1]}}\right) (n[l−1]2)
def initialize_parameters_he(layers_dims):
"""
Arguments:
layer_dims -- python array (list) containing the size of each layer.
Returns:
parameters -- python dictionary containing your parameters "W1", "b1", ..., "WL", "bL":
W1 -- weight matrix of shape (layers_dims[1], layers_dims[0])
b1 -- bias vector of shape (layers_dims[1], 1)
...
WL -- weight matrix of shape (layers_dims[L], layers_dims[L-1])
bL -- bias vector of shape (layers_dims[L], 1)
"""
parameters = {}
L = len(layers_dims) - 1 # integer representing the number of layers
for l in range(1, L + 1):
parameters['W' + str(l)] = np.random.randn(layers_dims[l],layers_dims[l-1])*np.sqrt(2./layers_dims[l-1])
parameters['b' + str(l)] = np.zeros((layers_dims[l],1))
return parameters
3. Activation function(激活函数)
- sigmoid function
s i g m o i d ( z ) = 1 1 + e − z sigmoid(z) = \frac{1}{1+e^{-z}} sigmoid(z)=1+e−z1
def sigmoid(Z):
"""
Implements the sigmoid activation in numpy
Arguments:
Z -- numpy array of any shape
Returns:
A -- output of sigmoid(z), same shape as Z
cache -- returns Z as well, useful during backpropagation
"""
A = 1/(1+np.exp(-Z))
cache_Z = Z
return A, cache_Z
- sigmoid backward function
s i g m o i d _ d e r i v a t i v e ( z ) = σ ′ ( z ) = σ ( z ) ( 1 − σ ( z ) ) sigmoid\_derivative(z) = \sigma'(z) = \sigma(z) (1 - \sigma(z)) sigmoid_derivative(z)=σ′(z)=σ(z)(1−σ(z))
def sigmoid_backward(dA, cache):
"""
Implement the backward propagation for a single SIGMOID unit.
Arguments:
dA -- post-activation gradient, of any shape
cache -- 'Z' where we store for computing backward propagation efficiently
Returns:
dZ -- Gradient of the cost with respect to Z
"""
Z = cache
s, cache_Z= sigmoid(Z)
dZ = dA * s * (1-s)
return dZ
- relu function
g ( z ) = max ( 0 , z ) g(z)=\max (0, z) g(z)=max(0,z)
def relu(Z):
"""
Implement the RELU function.
Arguments:
Z -- Output of the linear layer, of any shape
Returns:
A -- Post-activation parameter, of the same shape as Z
cache -- a python dictionary containing "A" ; stored for computing the backward pass efficiently
"""
A = np.maximum(0,Z)
cache_Z = Z
return A, cache_Z
- relu backward function
g ( z ) ′ = { 0 if z < 0 1 if z > 0 u n d e f i n e d if z = 0 g(z)^{\prime}=\left\{\begin{array}{ll}{0} & {\text { if } z<0} \\ {1} & {\text { if } z>0} \\ {u n d e f i n e d} & {\text { if } z=0}\end{array}\right. g(z)′=⎩⎨⎧01undefined if z<0 if z>0 if z=0
def relu_backward(dA, cache):
"""
Implement the backward propagation for a single RELU unit.
Arguments:
dA -- post-activation gradient, of any shape
cache -- 'Z' where we store for computing backward propagation efficiently
Returns:
dZ -- Gradient of the cost with respect to Z
"""
Z = cache
dZ = np.array(dA, copy=True) # just converting dz to a correct object.
# When z <= 0, you should set dz to 0 as well.
dZ[Z <= 0] = 0
return dZ
4. Forward propagation(正向传播)
- activation function
Z [ l ] = W [ l ] A [ l − 1 ] + b [ l ] Z^{[l]} = W^{[l]}A^{[l-1]} +b^{[l]} Z[l]=W[l]A[l−1]+b[l]
def linear_forward(A, W, b):
"""
Implement the linear part of a layer's forward propagation.
Arguments:
A -- activations from previous layer (or input data): (size of previous layer, number of examples)
W -- weights matrix: numpy array of shape (size of current layer, size of previous layer)
b -- bias vector, numpy array of shape (size of the current layer, 1)
Returns:
Z -- the input of the activation function, also called pre-activation parameter
cache -- a python dictionary containing "A", "W" and "b" ; stored for computing the backward pass efficiently
"""
Z = W.dot(A) + b
cache_AWb = (A, W, b)
return Z, cache_AWb
def linear_activation_forward(A_prev, W, b, activation):
"""
Implement the forward propagation for the LINEAR->ACTIVATION layer
Arguments:
A_prev -- activations from previous layer (or input data): (size of previous layer, number of examples)
W -- weights matrix: numpy array of shape (size of current layer, size of previous layer)
b -- bias vector, numpy array of shape (size of the current layer, 1)
activation -- the activation to be used in this layer, stored as a text string: "sigmoid" or "relu"
Returns:
A -- the output of the activation function, also called the post-activation value
cache -- a python dictionary containing "linear_cache" and "activation_cache";
stored for computing the backward pass efficiently
"""
if activation == "sigmoid":
# Inputs: "A_prev, W, b". Outputs: "A, activation_cache".
Z, linear_cache_AWb = linear_forward(A_prev, W, b)
A, activation_cache_Z = sigmoid(Z)
elif activation == "relu":
# Inputs: "A_prev, W, b". Outputs: "A, activation_cache".
Z, linear_cache_AWb = linear_forward(A_prev, W, b)
A, activation_cache_Z = relu(Z)
cache_AWbZ = (linear_cache_AWb, activation_cache_Z)
'''cache_AWbZ = ((Al-1, Wl, bl), Zl)'''
return A, cache_AWbZ
- L_model_forward
A [ l ] = g ( Z [ l ] ) = g ( W [ l ] A [ l − 1 ] + b [ l ] ) A^{[l]} = g(Z^{[l]}) = g(W^{[l]}A^{[l-1]} +b^{[l]}) A[l]=g(Z[l])=g(W[l]A[l−1]+b[l])
def L_model_forward(X, parameters):
"""
Implement forward propagation for the [LINEAR->RELU]*(L-1)->LINEAR->SIGMOID computation
Arguments:
X -- data, numpy array of shape (input size, number of examples)
parameters -- output of initialize_parameters_deep()
Returns:
AL -- last post-activation value
caches -- list of caches containing:
every cache of linear_relu_forward() (there are L-1 of them, indexed from 0 to L-2)
the cache of linear_sigmoid_forward() (there is one, indexed L-1)
"""
caches = []
A = X
L = len(parameters) // 2 # number of layers in the neural network
# Implement [LINEAR -> RELU]*(L-1). Add "cache" to the "caches" list.
for l in range(1, L):
A_prev = A
'''cache_AWbZ = ((Al-1, Wl, bl), Zl)'''
A, cache_AWbZ = linear_activation_forward(A_prev, parameters['W' + str(l)], parameters['b' + str(l)], activation = "relu")
'''caches = [ ((A0/X, W1, b1), Z1), ((A1, W2, b2), Z2), ......, ((AL-2, WL-1, bL-1), ZL-1) ]'''
caches.append(cache_AWbZ)
# Implement LINEAR -> SIGMOID. Add "cache" to the "caches" list.
'''cache_AWbZ = ((Al-1, Wl, bl), Zl)'''
AL, cache_AWbZ = linear_activation_forward(A, parameters['W' + str(L)], parameters['b' + str(L)], activation = "sigmoid")
'''caches = [ ((A0/X, W1, b1), Z1), ((A1, W2, b2), Z2), ......, ((AL-1, WL, bL), ZL) ]'''
caches.append(cache_AWbZ)
'''caches = [ ((A0/X, W1, b1), Z1), ((A1, W2, b2), Z2), ......, ((AL-1, WL, bL), ZL) ]'''
return AL, caches
- Forward propagation with dropout
def L_model_forward_with_dropout(X, parameters, keep_prob = 0.5):
"""
Implements the forward propagation: LINEAR -> RELU + DROPOUT -> LINEAR -> RELU + DROPOUT -> LINEAR -> SIGMOID.
Arguments:
X -- input dataset, of shape (2, number of examples)
parameters -- python dictionary containing your parameters "W", "b"
keep_prob - probability of keeping a neuron active during drop-out, scalar
Returns:
AL -- last activation value, output of the forward propagation
cache -- tuple, information stored for computing the backward propagation
"""
caches = []
A = X
L = len(parameters) // 2 # number of layers in the neural network
# [LINEAR -> RELU] × (L-1) -> LINEAR -> SIGMOID backward (whole model)
for l in range(1, L):
A_prev = A
D = np.random.rand(A.shape[0],A.shape[1]) # Step 1: initialize matrix D1 = np.random.rand(..., ...)
D = D < keep_prob # Step 2: convert entries of D1 to 0 or 1 (using keep_prob as the threshold)
A_prev = A_prev * D # Step 3: shut down some neurons of A1
A_prev = A_prev / keep_prob # Step 4: scale the value of neurons that haven't been shut down
'''cache_AWbZ = ((A, W, b), Z)'''
A, cache_AWbZ = linear_activation_forward(A_prev, parameters['W' + str(l)], parameters['b' + str(l)], activation = "relu")
'''caches = [ ((A1, W1, b1), Z1), ((A2, W2, b2), Z2), ......, ((AL-1, WL-1, bL-1), ZL-1) ]'''
caches.append(cache_AWbZ)
# Implement LINEAR -> SIGMOID. Add "cache" to the "caches" list.
'''cache_AWbZ = ((A, W, b), Z)'''
AL, cache_AWbZ = linear_activation_forward(A, parameters['W' + str(L)], parameters['b' + str(L)], activation = "sigmoid")
'''caches = [ ((A1, W1, b1), Z1), ((A2, W2, b2), Z2), ......, ((AL, WL, bL), ZL) ]'''
caches.append(cache_AWbZ)
'''caches = [ ((A1, W1, b1), Z1), ((A2, W2, b2), Z2), ......, ((AL, WL, bL), ZL) ]'''
return AL, cache
5. Cost function(代价函数)
- compute cost
J = − 1 m ∑ i = 1 m ( y ( i ) log ( a [ L ] ( i ) ) + ( 1 − y ( i ) ) log ( 1 − a [ L ] ( i ) ) ) J = -\frac{1}{m} \sum\limits_{i = 1}^{m} \large{(}\small y^{(i)}\log\left(a^{[L](i)}\right) + (1-y^{(i)})\log\left(1- a^{[L](i)}\right) \large{)} J=−m1i=1∑m(y(i)log(a[L](i))+(1−y(i))log(1−a[L](i)))
def compute_cost(AL, Y):
"""
Implement the cost function defined by equation (7).
Arguments:
AL -- probability vector corresponding to your label predictions, shape (1, number of examples)
Y -- true "label" vector (for example: containing 0 if non-cat, 1 if cat), shape (1, number of examples)
Returns:
cost -- cross-entropy cost
"""
m = Y.shape[1]
# Compute loss from aL and y.
cost = -1 / m * np.sum(Y * np.log(AL) + (1-Y) * np.log(1-AL),axis=1,keepdims=True)
cost = np.squeeze(cost) # To make sure your cost's shape is what we expect (e.g. this turns [[17]] into 17).
return cost
- compute cost with regularization
J r e g u l a r i z e d = − 1 m ∑ i = 1 m ( y ( i ) log ( a [ L ] ( i ) ) + ( 1 − y ( i ) ) log ( 1 − a [ L ] ( i ) ) ) ⎵ cross-entropy cost + 1 m λ 2 ∑ l ∑ k ∑ j W k , j [ l ] 2 ⎵ L2 regularization cost J_{regularized} = \small \underbrace{-\frac{1}{m} \sum\limits_{i = 1}^{m} \large{(}\small y^{(i)}\log\left(a^{[L](i)}\right) + (1-y^{(i)})\log\left(1- a^{[L](i)}\right) \large{)} }_\text{cross-entropy cost} + \underbrace{\frac{1}{m} \frac{\lambda}{2} \sum\limits_l\sum\limits_k\sum\limits_j W_{k,j}^{[l]2} }_\text{L2 regularization cost} Jregularized=cross-entropy cost −m1i=1∑m(y(i)log(a[L](i))+(1−y(i))log(1−a[L](i)))+L2 regularization cost m12λl∑k∑j∑Wk,j[l]2
def compute_cost_with_regularization(AL, Y, parameters, lambd):
"""
Implement the cost function with L2 regularization. See formula (2) above.
Arguments:
A3 -- post-activation, output of forward propagation, of shape (output size, number of examples)
Y -- "true" labels vector, of shape (output size, number of examples)
parameters -- python dictionary containing parameters of the model
Returns:
cost - value of the regularized loss function (formula (2))
"""
m = Y.shape[1]
L = len(parameters)//2
cross_entropy_cost = compute_cost(AL, Y) # This gives you the cross-entropy part of the cost
L2_regularization_cost = 0
for l in range(1, L):
L2_regularization_cost += np.sum(np.square(parameters['W' + str(l)]))
cost = cross_entropy_cost + (1./m*lambd/2)*(L2_regularization_cost)
return cost
- linear backward function with regularization
d W [ l ] = ∂ L ∂ W [ l ] = 1 m d Z [ l ] A [ l − 1 ] T + λ m ∗ W [ l ] dW^{[l]} = \frac{\partial \mathcal{L} }{\partial W^{[l]}} = \frac{1}{m} dZ^{[l]} A^{[l-1] T} +\frac{\lambda}{m}*W^{[l]} dW[l]=∂W[l]∂L=m1dZ[l]A[l−1]T+mλ∗W[l] d b [ l ] = ∂ L ∂ b [ l ] = 1 m ∑ i = 1 m d Z [ l ] ( i ) db^{[l]} = \frac{\partial \mathcal{L} }{\partial b^{[l]}} = \frac{1}{m} \sum_{i = 1}^{m} dZ^{[l](i)} db[l]=∂b[l]∂L=m1i=1∑mdZ[l](i) d A [ l − 1 ] = ∂ L ∂ A [ l − 1 ] = W [ l ] T d Z [ l ] dA^{[l-1]} = \frac{\partial \mathcal{L} }{\partial A^{[l-1]}} = W^{[l] T} dZ^{[l]} dA[l−1]=∂A[l−1]∂L=W[l]TdZ[l]
def linear_backward(dZ, linear_cache, lambd):
"""
Implement the linear portion of backward propagation for a single layer (layer l)
Arguments:
dZ -- Gradient of the cost with respect to the linear output (of current layer l)
linear_cache -- tuple of values (A_prev, W, b) coming from the forward propagation in the current layer
'''linear_cache = (AL, WL, bL)'''
Returns:
dA_prev -- Gradient of the cost with respect to the activation (of the previous layer l-1), same shape as A_prev
dW -- Gradient of the cost with respect to W (current layer l), same shape as W
db -- Gradient of the cost with respect to b (current layer l), same shape as b
"""
A_prev, W, b = linear_cache
m = A_prev.shape[1]
dW = 1./m * np.dot(dZ, A_prev.T) + lambd/m * W
db = 1./m * np.sum(dZ, axis=1, keepdims = True)
dA_prev = np.dot(W.T, dZ)
return dA_prev, dW, db
def linear_activation_backward(dA, cache, lambd, activation):
"""
Implement the backward propagation for the LINEAR->ACTIVATION layer.
Arguments:
dA -- post-activation gradient for current layer l
cache -- tuple of values (linear_cache, activation_cache) we store for computing backward propagation efficiently
'''current_cache = ((AL, WL, bL), ZL)'''
activation -- the activation to be used in this layer, stored as a text string: "sigmoid" or "relu"
Returns:
dA_prev -- Gradient of the cost with respect to the activation (of the previous layer l-1), same shape as A_prev
dW -- Gradient of the cost with respect to W (current layer l), same shape as W
db -- Gradient of the cost with respect to b (current layer l), same shape as b
"""
linear_cache, activation_cache = cache
'''linear_cache = (Al-1, Wl, bl), activation_cache= Zl '''
if activation == "relu":
dZ = relu_backward(dA, activation_cache)
dA_prev, dW, db = linear_backward(dZ, linear_cache, lambd)
elif activation == "sigmoid":
dZ = sigmoid_backward(dA, activation_cache)
dA_prev, dW, db = linear_backward(dZ, linear_cache, lambd)
return dA_prev, dW, db
backward propagation with regularization
def L_model_backward_with_regularization(AL, Y, caches, lambd):
"""
Implements the backward propagation of our baseline model to which we added an L2 regularization.
Arguments:
X -- input dataset, of shape (input size, number of examples)
Y -- "true" labels vector, of shape (output size, number of examples)
caches -- list of caches containing:
'''caches = [ ((A0/X, W1, b1), Z1), ((A1, W2, b2), Z2), ......, ((AL-1, WL, bL), ZL) ]'''
every cache of linear_activation_forward() with "relu" (there are (L-1) or them, indexes from 0 to L-2)
the cache of linear_activation_forward() with "sigmoid" (there is one, index L-1)
lambd -- regularization hyperparameter, scalar
Returns:
gradients -- A dictionary with the gradients with respect to each parameter, activation and pre-activation variables
"""
grads = {}
L = len(caches) # the number of layers
m = AL.shape[1]
Y = Y.reshape(AL.shape) # after this line, Y is the same shape as AL
# Initializing the backpropagation
dAL = - (np.divide(Y, AL) - np.divide(1 - Y, 1 - AL))
# Lth layer (SIGMOID -> LINEAR) gradients. Inputs: "AL, Y, caches". Outputs: "grads["dAL"], grads["dWL"], grads["dbL"]
'''current_cache = ((AL-1, WL, bL), ZL)'''
current_cache = caches[L-1]
grads["dA" + str(L)], grads["dW" + str(L)], grads["db" + str(L)] = linear_activation_backward(dAL, current_cache, lambd, activation = "sigmoid")
for l in reversed(range(L-1)):
''' l = L-2, L-3, ...., 2, 1, 0 '''
# lth layer: (RELU -> LINEAR) gradients.
'''current_cache = (AL-2, WL-1, bL-1), ZL-1)'''
current_cache = caches[l]
dA_prev_temp, dW_temp, db_temp = linear_activation_backward(grads["dA" + str(l + 2)], current_cache, lambd, activation = "relu")
grads["dA" + str(l + 1)] = dA_prev_temp
grads["dW" + str(l + 1)] = dW_temp
grads["db" + str(l + 1)] = db_temp
return grads
6. Update parameters(更新参数)
- update_parameters function
W [ l ] = W [ l ] − α d W [ l ] W^{[l]} = W^{[l]} - \alpha \text{ } dW^{[l]} W[l]=W[l]−α dW[l] b [ l ] = b [ l ] − α d b [ l ] b^{[l]} = b^{[l]} - \alpha \text{ } db^{[l]} b[l]=b[l]−α db[l]
def update_parameters(parameters, grads, learning_rate):
"""
Update parameters using gradient descent
Arguments:
parameters -- python dictionary containing your parameters
grads -- python dictionary containing your gradients, output of L_model_backward
Returns:
parameters -- python dictionary containing your updated parameters
parameters["W" + str(l)] = ...
parameters["b" + str(l)] = ...
"""
L = len(parameters) // 2 # number of layers in the neural network
# Update rule for each parameter. Use a for loop.
for l in range(L):
parameters["W" + str(l+1)] = parameters["W" + str(l+1)] - learning_rate * grads["dW" + str(l+1)]
parameters["b" + str(l+1)] = parameters["b" + str(l+1)] - learning_rate * grads["db" + str(l+1)]
return parameters
7. predict(预测结果)
def predict(X, y, parameters):
"""
This function is used to predict the results of a L-layer neural network.
Arguments:
X -- data set of examples you would like to label
parameters -- parameters of the trained model
Returns:
p -- predictions for the given dataset X
"""
m = X.shape[1]
n = len(parameters) // 2 # number of layers in the neural network
p = np.zeros((1,m))
# Forward propagation
probas, caches = L_model_forward(X, parameters)
# convert probas to 0/1 predictions
for i in range(0, probas.shape[1]):
if probas[0,i] > 0.5:
p[0,i] = 1
else:
p[0,i] = 0
print("Accuracy: " + str(np.sum((p == y)/m)))
return p
8.DNN
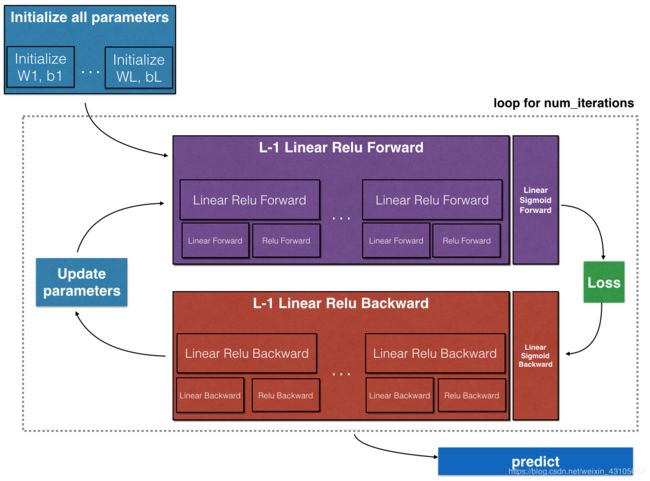
def L_layer_model(X, Y, layers_dims, learning_rate = 0.0075, num_iterations = 300, lambd = 0, print_cost=False, keep_prob = 0.5):#lr was 0.009
"""
Implements a L-layer neural network: [LINEAR->RELU]*(L-1)->LINEAR->SIGMOID.
Arguments:
X -- data, numpy array of shape (number of examples, num_px * num_px * 3)
Y -- true "label" vector (containing 0 if cat, 1 if non-cat), of shape (1, number of examples)
layers_dims -- list containing the input size and each layer size, of length (number of layers + 1).
learning_rate -- learning rate of the gradient descent update rule
num_iterations -- number of iterations of the optimization loop
print_cost -- if True, it prints the cost every 100 steps
Returns:
parameters -- parameters learnt by the model. They can then be used to predict.
"""
costs = [] # keep track of cost
# Parameters initialization.
parameters = initialize_parameters_random(layers_dims)
# Loop (gradient descent)
for i in range(0, num_iterations):
# Forward propagation: [LINEAR -> RELU]*(L-1) -> LINEAR -> SIGMOID.
'''caches = [ ((A0, W1, b1), Z1), ((A1, W2, b2), Z2), ......, ((AL-1, WL, bL), ZL) ]'''
AL, caches = L_model_forward(X, parameters)
'''caches = [ ((A0, W1, b1), Z1), ((A1, W2, b2), Z2), ......, ((AL-1, WL, bL), ZL) ]'''
# AL, caches = L_model_forward_with_dropout(X, parameters, keep_prob = 0.5)
# Compute cost.
# cost = compute_cost(AL, Y)
cost = compute_cost_with_regularization(AL, Y, parameters, lambd)
# Backward propagation.
'''caches = [ ((A0, W1, b1), Z1), ((A1, W2, b2), Z2), ......, ((AL-1, WL, bL), ZL) ]'''
grads = L_model_backward_with_regularization(AL, Y, caches, lambd)
# Update parameters.
parameters = update_parameters(parameters, grads, learning_rate)
# Print the cost every 100 training example
if print_cost and i % 100 == 0:
print ("Cost after iteration %i: %f" %(i, cost))
if print_cost and i % 100 == 0:
costs.append(cost)
# plot the cost
plt.plot(np.squeeze(costs))
plt.ylabel('cost')
plt.xlabel('iterations (per tens)')
plt.title("Learning rate =" + str(learning_rate))
plt.show()
return parameters


