Tensorflow 笔记 Ⅵ——Titanic Keras建模与应用
文章目录
- 数据集
- 基本原理
- Titanic TensorFLow 2.x Keras API 实现
- 前情函数介绍
- Pandas 缺失值判断函数 isnull()、isnull().any()、isnull().sum()
- Pandas 缺失值填充函数 fillna()
- Pandas map() 映射函数
- Pandas sample() 数据洗牌
- Pandas drop() 删除数据
- Pandas values 转换为 ndarray
- 正式开始
- 导入必要包
- 数据下载
- 读取数据
- 数据预处理
- 空值填充
- 数字映射
- 创建训练数据集与标签
- 数据标准化
- 代码重构定义数据预处理函数
- 数据读取并洗牌
- 训练可视化
- 测试模型
- 使用 Jack & Rose 测试
- 数据插入最后一列生存概率
- 加入回调
- 加载模型
- Titanic TensorFLow 1.x Keras API 实现
- 导入必要的包
- 定义预处理函数
- 读取数据,制作数据集
- 搭建模型
- 开始训练
- 训练可视化
- 模型预测
- 加载模型进行预测

数据集
数据集记录了泰坦尼克号一部分人的信息,以及其存活率等,众所周知,泰坦尼克号是一场海难,也就造成了人员信息难以调查,所以数据集中具有一些缺失的数据,数据集可以在 Kaggle 下载,也可以点击此处下载
| pclass | survived | name | sex | age | sibsp | parch | ticket | fare | cabin | embarked | boat | body | home.dest | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 1 | Allen, Miss. Elisabeth Walton | female | 29.0000 | 0 | 0 | 24160 | 211.3375 | B5 | S | 2 | NaN | St Louis, MO |
| 1 | 1 | 1 | Allison, Master. Hudson Trevor | male | 0.9167 | 1 | 2 | 113781 | 151.5500 | C22 C26 | S | 11 | NaN | Montreal, PQ / Chesterville, ON |
| 2 | 1 | 0 | Allison, Miss. Helen Loraine | female | 2.0000 | 1 | 2 | 113781 | 151.5500 | C22 C26 | S | NaN | NaN | Montreal, PQ / Chesterville, ON |
| 3 | 1 | 0 | Allison, Mr. Hudson Joshua Creighton | male | 30.0000 | 1 | 2 | 113781 | 151.5500 | C22 C26 | S | NaN | 135.0 | Montreal, PQ / Chesterville, ON |
| 4 | 1 | 0 | Allison, Mrs. Hudson J C (Bessie Waldo Daniels) | female | 25.0000 | 1 | 2 | 113781 | 151.5500 | C22 C26 | S | NaN | NaN | Montreal, PQ / Chesterville, ON |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1304 | 3 | 0 | Zabour, Miss. Hileni | female | 14.5000 | 1 | 0 | 2665 | 14.4542 | NaN | C | NaN | 328.0 | NaN |
| 1305 | 3 | 0 | Zabour, Miss. Thamine | female | NaN | 1 | 0 | 2665 | 14.4542 | NaN | C | NaN | NaN | NaN |
| 1306 | 3 | 0 | Zakarian, Mr. Mapriededer | male | 26.5000 | 0 | 0 | 2656 | 7.2250 | NaN | C | NaN | 304.0 | NaN |
| 1307 | 3 | 0 | Zakarian, Mr. Ortin | male | 27.0000 | 0 | 0 | 2670 | 7.2250 | NaN | C | NaN | NaN | NaN |
| 1308 | 3 | 0 | Zimmerman, Mr. Leo | male | 29.0000 | 0 | 0 | 315082 | 7.8750 | NaN | S | NaN | NaN | NaN |
1309 rows × 14 columns
泰坦尼克标签说明
| 字段 | 字段说明 | 数据说明 |
|---|---|---|
| pclass | 舱等级 | 1头等舱、2二等舱、3三等舱 |
| survival | 是否生存 | 0否、1是 |
| name | 姓名 | None |
| sex | 性别 | Female女性、male男性 |
| age | 年龄 | None |
| sibsp | siblings + parents | 兄弟姐妹或父母是否同船 |
| parch | parents + children | parents父母、children孩子 |
| ticked | 船票号码 | None |
| fare | 船票费用 | None |
| cabin | 舱位号码 | None |
| embarked | 登船港口 | C=Cherbourg,Q=Queenstown,S=Southampton |
| home.dest | 家、目的地 | home、destination |
基本原理
利用了多元线性回归问题的预处理方式,在最后一层输出层划分为分类问题,有关多元线性回归与逻辑回归问题的原理,屈尊移驾这里
和那里,由于此问题属于逻辑回归中的二分类问题,所以激活函数选用了 Sigmod 函数,在上诉的“那里”连接中解释了 Sigmod 函数
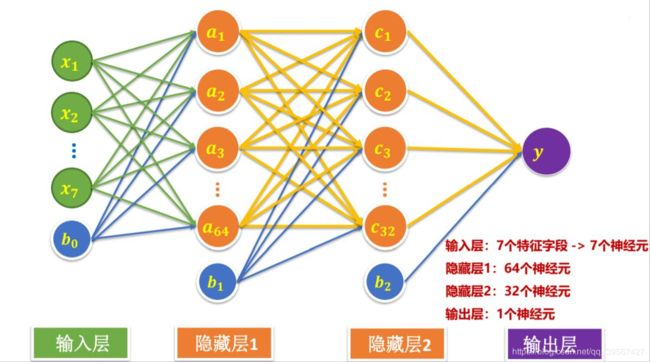
Titanic TensorFLow 2.x Keras API 实现
前情函数介绍
在 data 文件夹下,新建一个 xls 文件,内容如下
| int | char | float | |
|---|---|---|---|
| 0 | 1 | NaN | 1.1 |
| 1 | 2 | NaN | 2.2 |
| 2 | 3 | NaN | 3.3 |
| 3 | 4 | d | NaN |
| 4 | 5 | e | NaN |
| 5 | 6 | NaN | 6.6 |
| 6 | 7 | NaN | 7.7 |
import pandas as pd
demo_data = pd.read_excel('./data/demo.xls')
demo_data
| int | char | float | |
|---|---|---|---|
| 0 | 1 | NaN | 1.1 |
| 1 | 2 | NaN | 2.2 |
| 2 | 3 | NaN | 3.3 |
| 3 | 4 | d | NaN |
| 4 | 5 | e | NaN |
| 5 | 6 | NaN | 6.6 |
| 6 | 7 | NaN | 7.7 |
Pandas 缺失值判断函数 isnull()、isnull().any()、isnull().sum()
isnull() 返回一个 bool 值的 dataframe
isnull().any() 判断特征列是否存在空值
isnull().sum() 获取特征列空值数目
demo_data.isnull()
| int | char | float | |
|---|---|---|---|
| 0 | False | True | False |
| 1 | False | True | False |
| 2 | False | True | False |
| 3 | False | False | True |
| 4 | False | False | True |
| 5 | False | True | False |
| 6 | False | True | False |
demo_data.isnull().any()
int False
char True
float True
dtype: bool
demo_data.isnull().sum()
int 0
char 5
float 2
dtype: int64
Pandas 缺失值填充函数 fillna()
demo_data['char'] = demo_data['char'].fillna('A')
demo_data['float'] = demo_data['float'].fillna('5.5')
demo_data
| int | char | float | |
|---|---|---|---|
| 0 | 1 | A | 1.1 |
| 1 | 2 | A | 2.2 |
| 2 | 3 | A | 3.3 |
| 3 | 4 | d | 5.5 |
| 4 | 5 | e | 5.5 |
| 5 | 6 | A | 6.6 |
| 6 | 7 | A | 7.7 |
Pandas map() 映射函数
map 里面放入字典参数,将对应的键替换成其值,注意 map 需要将所有值全部替换,否则会报错
try:
demo_data['char'] = demo_data['char'].map({'A': 0, 'd': 1}).astype(int)
except ValueError:
print('Error rising:Cannot convert non-finite values (NA or inf) to integer')
finally:
demo_data['char'] = demo_data['char'].map({'A': 0, 'd': 1, 'e': 2}).astype(int)
print('Mapping finished')
Error rising:Cannot convert non-finite values (NA or inf) to integer
Mapping finished
demo_data
| int | char | float | |
|---|---|---|---|
| 0 | 1 | 0 | 1.1 |
| 1 | 2 | 0 | 2.2 |
| 2 | 3 | 0 | 3.3 |
| 3 | 4 | 1 | 5.5 |
| 4 | 5 | 2 | 5.5 |
| 5 | 6 | 0 | 6.6 |
| 6 | 7 | 0 | 7.7 |
Pandas sample() 数据洗牌
sample 用于在原数据中提取数据,并进行洗牌操作,frac 代表提取的比例,为 1 表示 100 % 100% 100%
shuffle_data_1 = demo_data.sample(frac = 1)
shuffle_data_1
| int | char | float | |
|---|---|---|---|
| 1 | 2 | 0 | 2.2 |
| 0 | 1 | 0 | 1.1 |
| 2 | 3 | 0 | 3.3 |
| 3 | 4 | 1 | 5.5 |
| 5 | 6 | 0 | 6.6 |
| 4 | 5 | 2 | 5.5 |
| 6 | 7 | 0 | 7.7 |
shuffle_data_2 = demo_data.sample(frac = 5 / 7)
shuffle_data_2
| int | char | float | |
|---|---|---|---|
| 6 | 7 | 0 | 7.7 |
| 0 | 1 | 0 | 1.1 |
| 2 | 3 | 0 | 3.3 |
| 5 | 6 | 0 | 6.6 |
| 1 | 2 | 0 | 2.2 |
Pandas drop() 删除数据
drop() 不改变原有数据,返回另一个 dataframe,使用 axis 可以指定行、列
demo_data = demo_data.drop(['char'], axis=1)
demo_data
| int | float | |
|---|---|---|
| 0 | 1 | 1.1 |
| 1 | 2 | 2.2 |
| 2 | 3 | 3.3 |
| 3 | 4 | 5.5 |
| 4 | 5 | 5.5 |
| 5 | 6 | 6.6 |
| 6 | 7 | 7.7 |
Pandas values 转换为 ndarray
经过上面处理,demo_data 中 data_frame 全部变成数字,使用 .values 将 data_frame 转换为 ndarray
nd_array = demo_data.values
print('nd_array:\n', nd_array,
'\nnd_array type:', type(nd_array))
nd_array:
[[1 1.1]
[2 2.2]
[3 3.3]
[4 '5.5']
[5 '5.5']
[6 6.6]
[7 7.7]]
nd_array type:
正式开始
导入必要包
import numpy
import pandas as pd
import tensorflow as tf
import urllib.request
from sklearn import preprocessing
import matplotlib.pyplot as plt
import os
import datetime
tf.__version__
'2.0.0'
数据下载
data_url = 'http://biostat.mc.vanderbilt.edu/wiki/pub/Main/DataSets/titanic3.xls'
data_file = './data/titanic3.xls'
if not os.path.exists(data_file):
operation = urllib.request.urlretrieve(data_url, data_file)
print('downloading from %s' % data_url)
else:
print('titanic3.xls is exists in the data directory')
titanic3.xls is exists in the data directory
读取数据
从数据摘要中发现 count 行的每一列数据不等,说明数据具有缺失项
dataframe = pd.read_excel(data_file)
dataframe.describe()
| pclass | survived | age | sibsp | parch | fare | body | |
|---|---|---|---|---|---|---|---|
| count | 1309.000000 | 1309.000000 | 1046.000000 | 1309.000000 | 1309.000000 | 1308.000000 | 121.000000 |
| mean | 2.294882 | 0.381971 | 29.881135 | 0.498854 | 0.385027 | 33.295479 | 160.809917 |
| std | 0.837836 | 0.486055 | 14.413500 | 1.041658 | 0.865560 | 51.758668 | 97.696922 |
| min | 1.000000 | 0.000000 | 0.166700 | 0.000000 | 0.000000 | 0.000000 | 1.000000 |
| 25% | 2.000000 | 0.000000 | 21.000000 | 0.000000 | 0.000000 | 7.895800 | 72.000000 |
| 50% | 3.000000 | 0.000000 | 28.000000 | 0.000000 | 0.000000 | 14.454200 | 155.000000 |
| 75% | 3.000000 | 1.000000 | 39.000000 | 1.000000 | 0.000000 | 31.275000 | 256.000000 |
| max | 3.000000 | 1.000000 | 80.000000 | 8.000000 | 9.000000 | 512.329200 | 328.000000 |
dataframe
| pclass | survived | name | sex | age | sibsp | parch | ticket | fare | cabin | embarked | boat | body | home.dest | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 1 | Allen, Miss. Elisabeth Walton | female | 29.0000 | 0 | 0 | 24160 | 211.3375 | B5 | S | 2 | NaN | St Louis, MO |
| 1 | 1 | 1 | Allison, Master. Hudson Trevor | male | 0.9167 | 1 | 2 | 113781 | 151.5500 | C22 C26 | S | 11 | NaN | Montreal, PQ / Chesterville, ON |
| 2 | 1 | 0 | Allison, Miss. Helen Loraine | female | 2.0000 | 1 | 2 | 113781 | 151.5500 | C22 C26 | S | NaN | NaN | Montreal, PQ / Chesterville, ON |
| 3 | 1 | 0 | Allison, Mr. Hudson Joshua Creighton | male | 30.0000 | 1 | 2 | 113781 | 151.5500 | C22 C26 | S | NaN | 135.0 | Montreal, PQ / Chesterville, ON |
| 4 | 1 | 0 | Allison, Mrs. Hudson J C (Bessie Waldo Daniels) | female | 25.0000 | 1 | 2 | 113781 | 151.5500 | C22 C26 | S | NaN | NaN | Montreal, PQ / Chesterville, ON |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1304 | 3 | 0 | Zabour, Miss. Hileni | female | 14.5000 | 1 | 0 | 2665 | 14.4542 | NaN | C | NaN | 328.0 | NaN |
| 1305 | 3 | 0 | Zabour, Miss. Thamine | female | NaN | 1 | 0 | 2665 | 14.4542 | NaN | C | NaN | NaN | NaN |
| 1306 | 3 | 0 | Zakarian, Mr. Mapriededer | male | 26.5000 | 0 | 0 | 2656 | 7.2250 | NaN | C | NaN | 304.0 | NaN |
| 1307 | 3 | 0 | Zakarian, Mr. Ortin | male | 27.0000 | 0 | 0 | 2670 | 7.2250 | NaN | C | NaN | NaN | NaN |
| 1308 | 3 | 0 | Zimmerman, Mr. Leo | male | 29.0000 | 0 | 0 | 315082 | 7.8750 | NaN | S | NaN | NaN | NaN |
1309 rows × 14 columns
数据预处理
去掉了 ticked、cabin,将 age、fare,空值用其列均值代替,sex 用0,1代替,embarked 用 S 代替,在将其所在字符值转为数字
注意:你应使用 .copy() 函数来防止严重警告⚠
selected_dataframe = dataframe[selected_cols].copy() ok
selected_dataframe = dataframe[selected_cols] not recommand
selected_cols = ['survived', 'name', 'pclass', 'sex', 'age', 'sibsp', 'parch', 'fare', 'embarked']
selected_dataframe = dataframe[selected_cols].copy()
空值填充
age_mean_value = selected_dataframe['age'].mean()
selected_dataframe['age'] = selected_dataframe['age'].fillna(age_mean_value)
fare_mean_value = selected_dataframe['fare'].mean()
selected_dataframe['fare'] = selected_dataframe['fare'].fillna(fare_mean_value)
selected_dataframe['embarked'] = selected_dataframe['embarked'].fillna('S')
selected_dataframe.describe()
| survived | pclass | age | sibsp | parch | fare | |
|---|---|---|---|---|---|---|
| count | 1309.000000 | 1309.000000 | 1309.000000 | 1309.000000 | 1309.000000 | 1309.000000 |
| mean | 0.381971 | 2.294882 | 29.881135 | 0.498854 | 0.385027 | 33.295479 |
| std | 0.486055 | 0.837836 | 12.883199 | 1.041658 | 0.865560 | 51.738879 |
| min | 0.000000 | 1.000000 | 0.166700 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 0.000000 | 2.000000 | 22.000000 | 0.000000 | 0.000000 | 7.895800 |
| 50% | 0.000000 | 3.000000 | 29.881135 | 0.000000 | 0.000000 | 14.454200 |
| 75% | 1.000000 | 3.000000 | 35.000000 | 1.000000 | 0.000000 | 31.275000 |
| max | 1.000000 | 3.000000 | 80.000000 | 8.000000 | 9.000000 | 512.329200 |
数字映射
selected_dataframe['sex'] = selected_dataframe['sex'].map({'female': 0, 'male': 1}).astype(int)
selected_dataframe['embarked'] = selected_dataframe['embarked'].map({'C': 0, 'Q': 1, 'S': 2}).astype(int)
创建训练数据集与标签
删除 name 列
selected_dataframe = selected_dataframe.drop(['name'], axis=1)
selected_dataframe[:3]
| survived | pclass | sex | age | sibsp | parch | fare | embarked | |
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 1 | 0 | 29.0000 | 0 | 0 | 211.3375 | 2 |
| 1 | 1 | 1 | 1 | 0.9167 | 1 | 2 | 151.5500 | 2 |
| 2 | 0 | 1 | 0 | 2.0000 | 1 | 2 | 151.5500 | 2 |
features 代表特征,第 1 到最后一列
label 代表标签,第 0 列
ndarray_data = selected_dataframe.values
features = ndarray_data[:, 1:]
label = ndarray_data[:, 0]
print('features:\n', features,
'\nlabel:', label)
features:
[[ 1. 0. 29. ... 0. 211.3375 2. ]
[ 1. 1. 0.9167 ... 2. 151.55 2. ]
[ 1. 0. 2. ... 2. 151.55 2. ]
...
[ 3. 1. 26.5 ... 0. 7.225 0. ]
[ 3. 1. 27. ... 0. 7.225 0. ]
[ 3. 1. 29. ... 0. 7.875 2. ]]
label: [1. 1. 0. ... 0. 0. 0.]
数据标准化
minmax_scale = preprocessing.MinMaxScaler(feature_range=(0, 1))
norm_features = minmax_scale.fit_transform(features)
print('norm_features:\n', norm_features,
'\nlabel:', label)
norm_features:
[[0. 0. 0.36116884 ... 0. 0.41250333 1. ]
[0. 1. 0.00939458 ... 0.22222222 0.2958059 1. ]
[0. 0. 0.0229641 ... 0.22222222 0.2958059 1. ]
...
[1. 1. 0.32985358 ... 0. 0.01410226 0. ]
[1. 1. 0.33611663 ... 0. 0.01410226 0. ]
[1. 1. 0.36116884 ... 0. 0.01537098 1. ]]
label: [1. 1. 0. ... 0. 0. 0.]
代码重构定义数据预处理函数
def prepare_data(df_data):
df = df_data.drop(['name'], axis=1)
age_mean = df['age'].mean()
df['age'] = df['age'].fillna(age_mean)
fare_mean = df['fare'].mean()
df['fare'] = df['fare'].fillna(fare_mean)
df['sex'] = df['sex'].map({'female':0, 'male':1}).astype(int)
df['embarked'] = df['embarked'].fillna('S')
df['embarked'] = df['embarked'].map({'C':0, 'Q':1, 'S':2}).astype(int)
ndarray_data = df.values
features = ndarray_data[:, 1:]
label = ndarray_data[:, 0]
minmax_scale = preprocessing.MinMaxScaler(feature_range=(0, 1))
norm_features = minmax_scale.fit_transform(features)
return norm_features, label
数据读取并洗牌
dataframe = pd.read_excel('./data/titanic3.xls')
selected_cols= ['survived', 'name', 'pclass', 'sex', 'age', 'sibsp', 'parch', 'fare', 'embarked']
selected_dataframe = dataframe[selected_cols].copy()
selected_dataframe = selected_dataframe.sample(frac=1)
x_data, y_data = prepare_data(selected_dataframe)
train_size = int(len(x_data) * 0.8)
x_train = x_data[:train_size]
y_train = y_data[:train_size]
x_test = x_data[train_size:]
y_test = y_data[train_size:]
model = tf.keras.models.Sequential([
tf.keras.layers.Dense(units=64,
input_dim=7,
use_bias=True,
kernel_initializer='uniform',
bias_initializer='zeros',
activation='relu'),
tf.keras.layers.Dropout(rate=0.3),
tf.keras.layers.Dense(units=32, activation='sigmoid'),
tf.keras.layers.Dropout(rate=0.3),
tf.keras.layers.Dense(units=1, activation='sigmoid')
])
model.summary()
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense (Dense) (None, 64) 512
_________________________________________________________________
dropout (Dropout) (None, 64) 0
_________________________________________________________________
dense_1 (Dense) (None, 32) 2080
_________________________________________________________________
dropout_1 (Dropout) (None, 32) 0
_________________________________________________________________
dense_2 (Dense) (None, 1) 33
=================================================================
Total params: 2,625
Trainable params: 2,625
Non-trainable params: 0
_________________________________________________________________
model.compile(optimizer=tf.keras.optimizers.Adam(0.003),
loss='binary_crossentropy',
metrics=['accuracy'])
train_history = model.fit(
x=x_train,
y=y_train,
validation_split=0.2,
epochs=100,
batch_size=40,
verbose=1)
Train on 837 samples, validate on 210 samples
Epoch 1/100
837/837 [==============================] - 2s 2ms/sample - loss: 0.6968 - accuracy: 0.5412 - val_loss: 0.5988 - val_accuracy: 0.6571
Epoch 2/100
837/837 [==============================] - 0s 147us/sample - loss: 0.6342 - accuracy: 0.6404 - val_loss: 0.5504 - val_accuracy: 0.7571
Epoch 3/100
837/837 [==============================] - 0s 153us/sample - loss: 0.5555 - accuracy: 0.7276 - val_loss: 0.4904 - val_accuracy: 0.8143
Epoch 4/100
837/837 [==============================] - 0s 148us/sample - loss: 0.5088 - accuracy: 0.7766 - val_loss: 0.4638 - val_accuracy: 0.8095
Epoch 5/100
837/837 [==============================] - 0s 137us/sample - loss: 0.4948 - accuracy: 0.7814 - val_loss: 0.4526 - val_accuracy: 0.8095
Epoch 6/100
837/837 [==============================] - 0s 124us/sample - loss: 0.4832 - accuracy: 0.7897 - val_loss: 0.4526 - val_accuracy: 0.8000
Epoch 7/100
837/837 [==============================] - 0s 140us/sample - loss: 0.4672 - accuracy: 0.7861 - val_loss: 0.4508 - val_accuracy: 0.8000
Epoch 8/100
837/837 [==============================] - 0s 142us/sample - loss: 0.4707 - accuracy: 0.7897 - val_loss: 0.4431 - val_accuracy: 0.8190
Epoch 9/100
837/837 [==============================] - 0s 117us/sample - loss: 0.4760 - accuracy: 0.8005 - val_loss: 0.4452 - val_accuracy: 0.8000
Epoch 10/100
837/837 [==============================] - 0s 121us/sample - loss: 0.4568 - accuracy: 0.8017 - val_loss: 0.4414 - val_accuracy: 0.7952
Epoch 11/100
837/837 [==============================] - 0s 132us/sample - loss: 0.4533 - accuracy: 0.8100 - val_loss: 0.4473 - val_accuracy: 0.7952
Epoch 12/100
837/837 [==============================] - 0s 122us/sample - loss: 0.4624 - accuracy: 0.7933 - val_loss: 0.4527 - val_accuracy: 0.8000
Epoch 13/100
837/837 [==============================] - 0s 119us/sample - loss: 0.4452 - accuracy: 0.8088 - val_loss: 0.4455 - val_accuracy: 0.8000
Epoch 14/100
837/837 [==============================] - 0s 122us/sample - loss: 0.4564 - accuracy: 0.7993 - val_loss: 0.4430 - val_accuracy: 0.7952
Epoch 15/100
837/837 [==============================] - 0s 161us/sample - loss: 0.4722 - accuracy: 0.8005 - val_loss: 0.4404 - val_accuracy: 0.8000
Epoch 16/100
837/837 [==============================] - 0s 141us/sample - loss: 0.4660 - accuracy: 0.8065 - val_loss: 0.4444 - val_accuracy: 0.8048
Epoch 17/100
837/837 [==============================] - 0s 124us/sample - loss: 0.4551 - accuracy: 0.8196 - val_loss: 0.4392 - val_accuracy: 0.8000
Epoch 18/100
837/837 [==============================] - 0s 118us/sample - loss: 0.4589 - accuracy: 0.8053 - val_loss: 0.4472 - val_accuracy: 0.8000
Epoch 19/100
837/837 [==============================] - 0s 116us/sample - loss: 0.4559 - accuracy: 0.8029 - val_loss: 0.4402 - val_accuracy: 0.8048
Epoch 20/100
837/837 [==============================] - 0s 122us/sample - loss: 0.4479 - accuracy: 0.8124 - val_loss: 0.4398 - val_accuracy: 0.7952
Epoch 21/100
837/837 [==============================] - 0s 122us/sample - loss: 0.4510 - accuracy: 0.8148 - val_loss: 0.4373 - val_accuracy: 0.8000
Epoch 22/100
837/837 [==============================] - 0s 126us/sample - loss: 0.4537 - accuracy: 0.8065 - val_loss: 0.4361 - val_accuracy: 0.8048
Epoch 23/100
837/837 [==============================] - 0s 119us/sample - loss: 0.4591 - accuracy: 0.8088 - val_loss: 0.4423 - val_accuracy: 0.7905
Epoch 24/100
837/837 [==============================] - 0s 119us/sample - loss: 0.4482 - accuracy: 0.8088 - val_loss: 0.4429 - val_accuracy: 0.7952
Epoch 25/100
837/837 [==============================] - 0s 121us/sample - loss: 0.4556 - accuracy: 0.8053 - val_loss: 0.4376 - val_accuracy: 0.8000
Epoch 26/100
837/837 [==============================] - 0s 128us/sample - loss: 0.4536 - accuracy: 0.8112 - val_loss: 0.4382 - val_accuracy: 0.8000
Epoch 27/100
837/837 [==============================] - 0s 119us/sample - loss: 0.4537 - accuracy: 0.7969 - val_loss: 0.4483 - val_accuracy: 0.8000
Epoch 28/100
837/837 [==============================] - 0s 120us/sample - loss: 0.4420 - accuracy: 0.8148 - val_loss: 0.4442 - val_accuracy: 0.7857
Epoch 29/100
837/837 [==============================] - 0s 125us/sample - loss: 0.4462 - accuracy: 0.8053 - val_loss: 0.4371 - val_accuracy: 0.7905
Epoch 30/100
837/837 [==============================] - 0s 125us/sample - loss: 0.4550 - accuracy: 0.8124 - val_loss: 0.4387 - val_accuracy: 0.7905
Epoch 31/100
837/837 [==============================] - 0s 139us/sample - loss: 0.4421 - accuracy: 0.8088 - val_loss: 0.4406 - val_accuracy: 0.7857
Epoch 32/100
837/837 [==============================] - 0s 122us/sample - loss: 0.4525 - accuracy: 0.8112 - val_loss: 0.4384 - val_accuracy: 0.7905
Epoch 33/100
837/837 [==============================] - 0s 126us/sample - loss: 0.4459 - accuracy: 0.8100 - val_loss: 0.4384 - val_accuracy: 0.7905
Epoch 34/100
837/837 [==============================] - 0s 133us/sample - loss: 0.4338 - accuracy: 0.8065 - val_loss: 0.4442 - val_accuracy: 0.7857
Epoch 35/100
837/837 [==============================] - 0s 143us/sample - loss: 0.4419 - accuracy: 0.8065 - val_loss: 0.4405 - val_accuracy: 0.7905
Epoch 36/100
837/837 [==============================] - 0s 137us/sample - loss: 0.4461 - accuracy: 0.8053 - val_loss: 0.4362 - val_accuracy: 0.7857
Epoch 37/100
837/837 [==============================] - 0s 118us/sample - loss: 0.4414 - accuracy: 0.8148 - val_loss: 0.4479 - val_accuracy: 0.7810
Epoch 38/100
837/837 [==============================] - 0s 120us/sample - loss: 0.4382 - accuracy: 0.8136 - val_loss: 0.4365 - val_accuracy: 0.7905
Epoch 39/100
837/837 [==============================] - 0s 125us/sample - loss: 0.4356 - accuracy: 0.8184 - val_loss: 0.4488 - val_accuracy: 0.7810
Epoch 40/100
837/837 [==============================] - 0s 143us/sample - loss: 0.4383 - accuracy: 0.8184 - val_loss: 0.4375 - val_accuracy: 0.7905
Epoch 41/100
837/837 [==============================] - 0s 150us/sample - loss: 0.4347 - accuracy: 0.8124 - val_loss: 0.4451 - val_accuracy: 0.7810
Epoch 42/100
837/837 [==============================] - 0s 143us/sample - loss: 0.4573 - accuracy: 0.8112 - val_loss: 0.4411 - val_accuracy: 0.7857
Epoch 43/100
837/837 [==============================] - 0s 119us/sample - loss: 0.4358 - accuracy: 0.8136 - val_loss: 0.4379 - val_accuracy: 0.7905
Epoch 44/100
837/837 [==============================] - 0s 164us/sample - loss: 0.4453 - accuracy: 0.8160 - val_loss: 0.4458 - val_accuracy: 0.7810
Epoch 45/100
837/837 [==============================] - 0s 126us/sample - loss: 0.4401 - accuracy: 0.8076 - val_loss: 0.4405 - val_accuracy: 0.7952
Epoch 46/100
837/837 [==============================] - 0s 142us/sample - loss: 0.4364 - accuracy: 0.8160 - val_loss: 0.4465 - val_accuracy: 0.7810
Epoch 47/100
837/837 [==============================] - 0s 129us/sample - loss: 0.4311 - accuracy: 0.8184 - val_loss: 0.4386 - val_accuracy: 0.8000
Epoch 48/100
837/837 [==============================] - 0s 117us/sample - loss: 0.4377 - accuracy: 0.8160 - val_loss: 0.4444 - val_accuracy: 0.7857
Epoch 49/100
837/837 [==============================] - 0s 133us/sample - loss: 0.4546 - accuracy: 0.7957 - val_loss: 0.4432 - val_accuracy: 0.7810
Epoch 50/100
837/837 [==============================] - 0s 145us/sample - loss: 0.4403 - accuracy: 0.8208 - val_loss: 0.4436 - val_accuracy: 0.7905
Epoch 51/100
837/837 [==============================] - 0s 144us/sample - loss: 0.4259 - accuracy: 0.8148 - val_loss: 0.4374 - val_accuracy: 0.7952
Epoch 52/100
837/837 [==============================] - 0s 155us/sample - loss: 0.4300 - accuracy: 0.8160 - val_loss: 0.4411 - val_accuracy: 0.7857
Epoch 53/100
837/837 [==============================] - 0s 160us/sample - loss: 0.4381 - accuracy: 0.8136 - val_loss: 0.4432 - val_accuracy: 0.7905
Epoch 54/100
837/837 [==============================] - 0s 136us/sample - loss: 0.4290 - accuracy: 0.8256 - val_loss: 0.4414 - val_accuracy: 0.7810
Epoch 55/100
837/837 [==============================] - 0s 148us/sample - loss: 0.4360 - accuracy: 0.8160 - val_loss: 0.4385 - val_accuracy: 0.7952
Epoch 56/100
837/837 [==============================] - 0s 114us/sample - loss: 0.4364 - accuracy: 0.8232 - val_loss: 0.4415 - val_accuracy: 0.7810
Epoch 57/100
837/837 [==============================] - 0s 127us/sample - loss: 0.4364 - accuracy: 0.8076 - val_loss: 0.4397 - val_accuracy: 0.7952
Epoch 58/100
837/837 [==============================] - 0s 122us/sample - loss: 0.4370 - accuracy: 0.8148 - val_loss: 0.4378 - val_accuracy: 0.7905
Epoch 59/100
837/837 [==============================] - 0s 139us/sample - loss: 0.4435 - accuracy: 0.8088 - val_loss: 0.4444 - val_accuracy: 0.7810
Epoch 60/100
837/837 [==============================] - 0s 122us/sample - loss: 0.4354 - accuracy: 0.8172 - val_loss: 0.4372 - val_accuracy: 0.7952
Epoch 61/100
837/837 [==============================] - 0s 137us/sample - loss: 0.4375 - accuracy: 0.8112 - val_loss: 0.4420 - val_accuracy: 0.7857
Epoch 62/100
837/837 [==============================] - 0s 135us/sample - loss: 0.4307 - accuracy: 0.8136 - val_loss: 0.4392 - val_accuracy: 0.7905
Epoch 63/100
837/837 [==============================] - 0s 158us/sample - loss: 0.4362 - accuracy: 0.8160 - val_loss: 0.4406 - val_accuracy: 0.7952
Epoch 64/100
837/837 [==============================] - 0s 124us/sample - loss: 0.4405 - accuracy: 0.8124 - val_loss: 0.4440 - val_accuracy: 0.7810
Epoch 65/100
837/837 [==============================] - 0s 119us/sample - loss: 0.4298 - accuracy: 0.8232 - val_loss: 0.4386 - val_accuracy: 0.8000
Epoch 66/100
837/837 [==============================] - 0s 108us/sample - loss: 0.4284 - accuracy: 0.8065 - val_loss: 0.4446 - val_accuracy: 0.7905
Epoch 67/100
837/837 [==============================] - 0s 129us/sample - loss: 0.4340 - accuracy: 0.8148 - val_loss: 0.4438 - val_accuracy: 0.7905
Epoch 68/100
837/837 [==============================] - 0s 114us/sample - loss: 0.4389 - accuracy: 0.8088 - val_loss: 0.4381 - val_accuracy: 0.8000
Epoch 69/100
837/837 [==============================] - 0s 124us/sample - loss: 0.4338 - accuracy: 0.8220 - val_loss: 0.4386 - val_accuracy: 0.8000
Epoch 70/100
837/837 [==============================] - 0s 115us/sample - loss: 0.4323 - accuracy: 0.8184 - val_loss: 0.4423 - val_accuracy: 0.7952
Epoch 71/100
837/837 [==============================] - 0s 122us/sample - loss: 0.4237 - accuracy: 0.8232 - val_loss: 0.4406 - val_accuracy: 0.7905
Epoch 72/100
837/837 [==============================] - 0s 137us/sample - loss: 0.4355 - accuracy: 0.8208 - val_loss: 0.4437 - val_accuracy: 0.7905
Epoch 73/100
837/837 [==============================] - 0s 116us/sample - loss: 0.4362 - accuracy: 0.8196 - val_loss: 0.4364 - val_accuracy: 0.7905
Epoch 74/100
837/837 [==============================] - 0s 128us/sample - loss: 0.4293 - accuracy: 0.8196 - val_loss: 0.4445 - val_accuracy: 0.7810
Epoch 75/100
837/837 [==============================] - 0s 124us/sample - loss: 0.4252 - accuracy: 0.8184 - val_loss: 0.4400 - val_accuracy: 0.7905
Epoch 76/100
837/837 [==============================] - 0s 119us/sample - loss: 0.4335 - accuracy: 0.8256 - val_loss: 0.4470 - val_accuracy: 0.7810
Epoch 77/100
837/837 [==============================] - 0s 119us/sample - loss: 0.4284 - accuracy: 0.8184 - val_loss: 0.4384 - val_accuracy: 0.8000
Epoch 78/100
837/837 [==============================] - 0s 147us/sample - loss: 0.4398 - accuracy: 0.8136 - val_loss: 0.4412 - val_accuracy: 0.7905
Epoch 79/100
837/837 [==============================] - 0s 129us/sample - loss: 0.4339 - accuracy: 0.8160 - val_loss: 0.4454 - val_accuracy: 0.7810
Epoch 80/100
837/837 [==============================] - 0s 127us/sample - loss: 0.4286 - accuracy: 0.8160 - val_loss: 0.4397 - val_accuracy: 0.7905
Epoch 81/100
837/837 [==============================] - 0s 120us/sample - loss: 0.4315 - accuracy: 0.8220 - val_loss: 0.4393 - val_accuracy: 0.7905
Epoch 82/100
837/837 [==============================] - 0s 138us/sample - loss: 0.4263 - accuracy: 0.8184 - val_loss: 0.4415 - val_accuracy: 0.7905
Epoch 83/100
837/837 [==============================] - 0s 136us/sample - loss: 0.4298 - accuracy: 0.8208 - val_loss: 0.4405 - val_accuracy: 0.8048
Epoch 84/100
837/837 [==============================] - 0s 129us/sample - loss: 0.4341 - accuracy: 0.8112 - val_loss: 0.4377 - val_accuracy: 0.7952
Epoch 85/100
837/837 [==============================] - 0s 127us/sample - loss: 0.4325 - accuracy: 0.8100 - val_loss: 0.4432 - val_accuracy: 0.8000
Epoch 86/100
837/837 [==============================] - 0s 132us/sample - loss: 0.4277 - accuracy: 0.8124 - val_loss: 0.4415 - val_accuracy: 0.7857
Epoch 87/100
837/837 [==============================] - 0s 113us/sample - loss: 0.4274 - accuracy: 0.8196 - val_loss: 0.4427 - val_accuracy: 0.7905
Epoch 88/100
837/837 [==============================] - 0s 112us/sample - loss: 0.4243 - accuracy: 0.8280 - val_loss: 0.4400 - val_accuracy: 0.7905
Epoch 89/100
837/837 [==============================] - 0s 118us/sample - loss: 0.4280 - accuracy: 0.8220 - val_loss: 0.4418 - val_accuracy: 0.7952
Epoch 90/100
837/837 [==============================] - 0s 112us/sample - loss: 0.4340 - accuracy: 0.8208 - val_loss: 0.4409 - val_accuracy: 0.8000
Epoch 91/100
837/837 [==============================] - 0s 121us/sample - loss: 0.4298 - accuracy: 0.8136 - val_loss: 0.4403 - val_accuracy: 0.8000
Epoch 92/100
837/837 [==============================] - 0s 122us/sample - loss: 0.4275 - accuracy: 0.8208 - val_loss: 0.4409 - val_accuracy: 0.7952
Epoch 93/100
837/837 [==============================] - 0s 136us/sample - loss: 0.4228 - accuracy: 0.8244 - val_loss: 0.4394 - val_accuracy: 0.8095
Epoch 94/100
837/837 [==============================] - 0s 119us/sample - loss: 0.4313 - accuracy: 0.8208 - val_loss: 0.4434 - val_accuracy: 0.8000
Epoch 95/100
837/837 [==============================] - 0s 130us/sample - loss: 0.4277 - accuracy: 0.8196 - val_loss: 0.4365 - val_accuracy: 0.8095
Epoch 96/100
837/837 [==============================] - 0s 118us/sample - loss: 0.4273 - accuracy: 0.8220 - val_loss: 0.4383 - val_accuracy: 0.8000
Epoch 97/100
837/837 [==============================] - 0s 113us/sample - loss: 0.4311 - accuracy: 0.8124 - val_loss: 0.4373 - val_accuracy: 0.8095
Epoch 98/100
837/837 [==============================] - 0s 123us/sample - loss: 0.4221 - accuracy: 0.8327 - val_loss: 0.4419 - val_accuracy: 0.7952
Epoch 99/100
837/837 [==============================] - 0s 124us/sample - loss: 0.4378 - accuracy: 0.8196 - val_loss: 0.4380 - val_accuracy: 0.8048
Epoch 100/100
837/837 [==============================] - 0s 122us/sample - loss: 0.4238 - accuracy: 0.8232 - val_loss: 0.4451 - val_accuracy: 0.7857
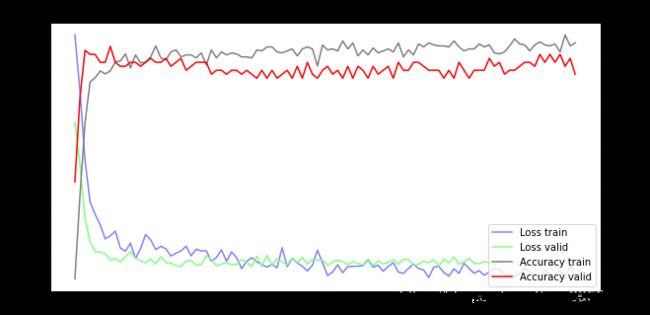
训练可视化
fig = plt.gcf()
fig.set_size_inches(10, 5)
ax1 = fig.add_subplot(111)
ax1.set_title('Train and Validation Picture')
ax1.set_ylabel('Loss value')
line1, = ax1.plot(train_history.history['loss'], color=(0.5, 0.5, 1.0), label='Loss train')
line2, = ax1.plot(train_history.history['val_loss'], color=(0.5, 1.0, 0.5), label='Loss valid')
ax2 = ax1.twinx()
ax2.set_ylabel('Accuracy value')
line3, = ax2.plot(train_history.history['accuracy'], color=(0.5, 0.5, 0.5), label='Accuracy train')
line4, = ax2.plot(train_history.history['val_accuracy'], color=(1, 0, 0), label='Accuracy valid')
plt.legend(handles=(line1, line2, line3, line4), loc='best')
plt.show()
测试模型
test_loss, test_acc = model.evaluate(x_test, y_test, verbose=2)
print('test_loss:', test_loss,
'\ntest_acc:', test_acc,
'\nmetrics_names:', model.metrics_names)
262/1 - 0s - loss: 0.3581 - accuracy: 0.7672
test_loss: 0.48995060536242624
test_acc: 0.76717556
metrics_names: ['loss', 'accuracy']
使用 Jack & Rose 测试
Jack_info = [0, 'Jack', 3, 'male', 23, 1, 0, 5.0000, 'S']
Rose_info = [1, 'Rose', 1, 'female', 20, 1, 0, 100.0000, 'S']
new_passenger_pd = pd.DataFrame([Jack_info, Rose_info], columns=selected_cols)
all_passenger_pd = selected_dataframe.append(new_passenger_pd)
pred = model.predict(prepare_data(all_passenger_pd)[0])
print('Rose survived probability:', pred[-1:][0][0],
'\nJack survived probability:', pred[-2:][0][0])
Rose survived probability: 0.96711206
Jack survived probability: 0.12514974
数据插入最后一列生存概率
all_passenger_pd.insert(len(all_passenger_pd.columns), 'surv_prob', pred)
all_passenger_pd
| survived | name | pclass | sex | age | sibsp | parch | fare | embarked | surv_prob | |
|---|---|---|---|---|---|---|---|---|---|---|
| 75 | 0 | Colley, Mr. Edward Pomeroy | 1 | male | 47.0 | 0 | 0 | 25.5875 | S | 0.221973 |
| 321 | 0 | Wright, Mr. George | 1 | male | 62.0 | 0 | 0 | 26.5500 | S | 0.194789 |
| 712 | 0 | Celotti, Mr. Francesco | 3 | male | 24.0 | 0 | 0 | 8.0500 | S | 0.130646 |
| 345 | 0 | Berriman, Mr. William John | 2 | male | 23.0 | 0 | 0 | 13.0000 | S | 0.211936 |
| 1298 | 0 | Wittevrongel, Mr. Camille | 3 | male | 36.0 | 0 | 0 | 9.5000 | S | 0.106074 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 10 | 0 | Astor, Col. John Jacob | 1 | male | 47.0 | 1 | 0 | 227.5250 | C | 0.213140 |
| 434 | 1 | Hart, Miss. Eva Miriam | 2 | female | 7.0 | 0 | 2 | 26.2500 | S | 0.881991 |
| 690 | 0 | Brobeck, Mr. Karl Rudolf | 3 | male | 22.0 | 0 | 0 | 7.7958 | S | 0.136104 |
| 0 | 0 | Jack | 3 | male | 23.0 | 1 | 0 | 5.0000 | S | 0.125150 |
| 1 | 1 | Rose | 1 | female | 20.0 | 1 | 0 | 100.0000 | S | 0.967112 |
1311 rows × 10 columns
form =pd.DataFrame(columns=[column for column in all_passenger_pd], data=all_passenger_pd)
form.to_excel('./data/result.xls', encoding='utf-8', index=None, header=True)
加入回调
def prepare_data(df_data):
df = df_data.drop(['name'], axis=1)
age_mean = df['age'].mean()
df['age'] = df['age'].fillna(age_mean)
fare_mean = df['fare'].mean()
df['fare'] = df['fare'].fillna(fare_mean)
df['sex'] = df['sex'].map({'female':0, 'male':1}).astype(int)
df['embarked'] = df['embarked'].fillna('S')
df['embarked'] = df['embarked'].map({'C':0, 'Q':1, 'S':2}).astype(int)
ndarray_data = df.values
features = ndarray_data[:, 1:]
label = ndarray_data[:, 0]
minmax_scale = preprocessing.MinMaxScaler(feature_range=(0, 1))
norm_features = minmax_scale.fit_transform(features)
return norm_features, label
dataframe = pd.read_excel('./data/titanic3.xls')
selected_cols= ['survived', 'name', 'pclass', 'sex', 'age', 'sibsp', 'parch', 'fare', 'embarked']
selected_dataframe = dataframe[selected_cols].copy()
selected_dataframe = selected_dataframe.sample(frac=1)
x_data, y_data = prepare_data(selected_dataframe)
train_size = int(len(x_data) * 0.8)
x_train = x_data[:train_size]
y_train = y_data[:train_size]
x_test = x_data[train_size:]
y_test = y_data[train_size:]
model = tf.keras.models.Sequential([
tf.keras.layers.Dense(units=256,
input_dim=7,
use_bias=True,
kernel_initializer='uniform',
bias_initializer='zeros',
activation='relu'),
tf.keras.layers.Dropout(rate=0.3),
tf.keras.layers.Dense(units=128, activation='sigmoid'),
tf.keras.layers.Dropout(rate=0.3),
tf.keras.layers.Dense(units=64, activation='sigmoid'),
tf.keras.layers.Dropout(rate=0.3),
tf.keras.layers.Dense(units=32, activation='sigmoid'),
tf.keras.layers.Dropout(rate=0.3),
tf.keras.layers.Dense(units=1, activation='sigmoid')
])
model.summary()
Model: "sequential_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense_3 (Dense) (None, 256) 2048
_________________________________________________________________
dropout_2 (Dropout) (None, 256) 0
_________________________________________________________________
dense_4 (Dense) (None, 128) 32896
_________________________________________________________________
dropout_3 (Dropout) (None, 128) 0
_________________________________________________________________
dense_5 (Dense) (None, 64) 8256
_________________________________________________________________
dropout_4 (Dropout) (None, 64) 0
_________________________________________________________________
dense_6 (Dense) (None, 32) 2080
_________________________________________________________________
dropout_5 (Dropout) (None, 32) 0
_________________________________________________________________
dense_7 (Dense) (None, 1) 33
=================================================================
Total params: 45,313
Trainable params: 45,313
Non-trainable params: 0
_________________________________________________________________
model.compile(optimizer=tf.keras.optimizers.Adam(0.003),
loss='binary_crossentropy',
metrics=['accuracy'])
log_dir = os.path.join(
'logs2.x',
'train',
'plugins',
'profile',
datetime.datetime.now().strftime('%Y-%m-%d_%H-%M-%S'))
checkpoint_path = './checkpoint2.x/Titanic.{epoch:02d}.ckpt'
if not os.path.exists('./checkpoint2.x'):
os.mkdir('./checkpoint2.x')
callbacks = [tf.keras.callbacks.TensorBoard(log_dir=log_dir,
histogram_freq=2),
tf.keras.callbacks.ModelCheckpoint(filepath=checkpoint_path,
save_weights_only=True,
verbose=1,
period=5)]
WARNING:tensorflow:`period` argument is deprecated. Please use `save_freq` to specify the frequency in number of samples seen.
train_history = model.fit(x=x_train, y=y_train,
validation_split=0.2,
epochs=100,
batch_size=40,
callbacks=callbacks,
verbose=1)
部分训练如下
Train on 837 samples, validate on 210 samples
Epoch 80/100
760/837 [==========================>...] - ETA: 0s - loss: 0.4277 - accuracy: 0.8132
Epoch 00080: saving model to ./checkpoint2.x/Titanic.80.h5
837/837 [==============================] - 0s 302us/sample - loss: 0.4353 - accuracy: 0.8112 - val_loss: 0.4639 - val_accuracy: 0.7810
Epoch 81/100
837/837 [==============================] - 0s 270us/sample - loss: 0.4455 - accuracy: 0.8017 - val_loss: 0.4768 - val_accuracy: 0.7810
Epoch 82/100
837/837 [==============================] - 0s 211us/sample - loss: 0.4376 - accuracy: 0.7993 - val_loss: 0.4654 - val_accuracy: 0.7905
Epoch 83/100
837/837 [==============================] - 0s 278us/sample - loss: 0.4377 - accuracy: 0.8065 - val_loss: 0.4703 - val_accuracy: 0.7810
Epoch 84/100
837/837 [==============================] - 0s 232us/sample - loss: 0.4368 - accuracy: 0.8160 - val_loss: 0.4631 - val_accuracy: 0.7952
Epoch 85/100
360/837 [===========>..................] - ETA: 0s - loss: 0.4669 - accuracy: 0.8056
Epoch 00085: saving model to ./checkpoint2.x/Titanic.85.h5
837/837 [==============================] - 0s 292us/sample - loss: 0.4437 - accuracy: 0.8124 - val_loss: 0.4627 - val_accuracy: 0.7810
Epoch 86/100
837/837 [==============================] - 0s 197us/sample - loss: 0.4365 - accuracy: 0.8017 - val_loss: 0.4686 - val_accuracy: 0.7905
Epoch 87/100
837/837 [==============================] - 0s 288us/sample - loss: 0.4500 - accuracy: 0.8148 - val_loss: 0.4689 - val_accuracy: 0.7857
Epoch 88/100
837/837 [==============================] - 0s 208us/sample - loss: 0.4356 - accuracy: 0.8029 - val_loss: 0.4794 - val_accuracy: 0.7905
Epoch 89/100
837/837 [==============================] - 0s 239us/sample - loss: 0.4283 - accuracy: 0.8148 - val_loss: 0.4621 - val_accuracy: 0.7857
Epoch 90/100
440/837 [==============>...............] - ETA: 0s - loss: 0.4083 - accuracy: 0.8295
Epoch 00090: saving model to ./checkpoint2.x/Titanic.90.h5
837/837 [==============================] - 0s 258us/sample - loss: 0.4359 - accuracy: 0.8172 - val_loss: 0.4736 - val_accuracy: 0.7905
Epoch 91/100
837/837 [==============================] - 0s 299us/sample - loss: 0.4365 - accuracy: 0.8053 - val_loss: 0.4658 - val_accuracy: 0.7905
Epoch 92/100
837/837 [==============================] - 0s 319us/sample - loss: 0.4376 - accuracy: 0.8148 - val_loss: 0.4696 - val_accuracy: 0.7905
Epoch 93/100
837/837 [==============================] - 0s 355us/sample - loss: 0.4375 - accuracy: 0.8005 - val_loss: 0.4698 - val_accuracy: 0.7952
Epoch 94/100
837/837 [==============================] - 0s 205us/sample - loss: 0.4384 - accuracy: 0.8005 - val_loss: 0.4682 - val_accuracy: 0.7905
Epoch 95/100
440/837 [==============>...............] - ETA: 0s - loss: 0.4514 - accuracy: 0.7909
Epoch 00095: saving model to ./checkpoint2.x/Titanic.95.h5
837/837 [==============================] - 0s 344us/sample - loss: 0.4392 - accuracy: 0.8005 - val_loss: 0.4620 - val_accuracy: 0.7952
Epoch 96/100
837/837 [==============================] - 0s 219us/sample - loss: 0.4347 - accuracy: 0.8053 - val_loss: 0.4643 - val_accuracy: 0.7857
Epoch 97/100
837/837 [==============================] - 0s 309us/sample - loss: 0.4410 - accuracy: 0.8005 - val_loss: 0.4772 - val_accuracy: 0.7905
Epoch 98/100
837/837 [==============================] - 0s 230us/sample - loss: 0.4325 - accuracy: 0.8076 - val_loss: 0.4629 - val_accuracy: 0.7857
Epoch 99/100
837/837 [==============================] - 0s 267us/sample - loss: 0.4308 - accuracy: 0.8005 - val_loss: 0.4658 - val_accuracy: 0.7857
Epoch 100/100
360/837 [===========>..................] - ETA: 0s - loss: 0.4338 - accuracy: 0.8139
Epoch 00100: saving model to ./checkpoint2.x/Titanic.100.h5
837/837 [==============================] - 0s 265us/sample - loss: 0.4314 - accuracy: 0.8124 - val_loss: 0.4623 - val_accuracy: 0.7857
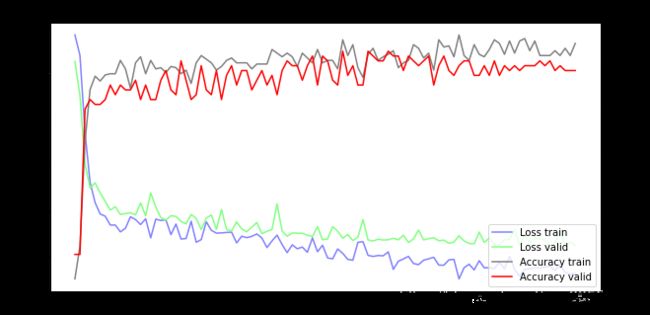
fig = plt.gcf()
fig.set_size_inches(10, 5)
ax1 = fig.add_subplot(111)
ax1.set_title('Train and Validation Picture')
ax1.set_ylabel('Loss value')
line1, = ax1.plot(train_history.history['loss'], color=(0.5, 0.5, 1.0), label='Loss train')
line2, = ax1.plot(train_history.history['val_loss'], color=(0.5, 1.0, 0.5), label='Loss valid')
ax2 = ax1.twinx()
ax2.set_ylabel('Accuracy value')
line3, = ax2.plot(train_history.history['accuracy'], color=(0.5, 0.5, 0.5), label='Accuracy train')
line4, = ax2.plot(train_history.history['val_accuracy'], color=(1, 0, 0), label='Accuracy valid')
plt.legend(handles=(line1, line2, line3, line4), loc='best')
plt.show()
Jack_info = [0, 'Jack', 3, 'male', 23, 1, 0, 5.0000, 'S']
Rose_info = [1, 'Rose', 1, 'female', 20, 1, 0, 100.0000, 'S']
new_passenger_pd = pd.DataFrame([Jack_info, Rose_info], columns=selected_cols)
all_passenger_pd = selected_dataframe.append(new_passenger_pd)
pred = model.predict(prepare_data(all_passenger_pd)[0])
print('Rose survived probability:', pred[-1:][0][0],
'\nJack survived probability:', pred[-2:][0][0])
Rose survived probability: 0.9700622
Jack survived probability: 0.12726058
加载模型
由于只保存了网络参数,没有保存网络结构,需要重新定义网络结构(当然,由于 jupyter 的缓存效应,你大可不必重新定义,对于独立的 py 文件则需要这么做)
model = tf.keras.models.Sequential([
tf.keras.layers.Dense(units=256,
input_dim=7,
use_bias=True,
kernel_initializer='uniform',
bias_initializer='zeros',
activation='relu'),
tf.keras.layers.Dropout(rate=0.3),
tf.keras.layers.Dense(units=128, activation='sigmoid'),
tf.keras.layers.Dropout(rate=0.3),
tf.keras.layers.Dense(units=64, activation='sigmoid'),
tf.keras.layers.Dropout(rate=0.3),
tf.keras.layers.Dense(units=32, activation='sigmoid'),
tf.keras.layers.Dropout(rate=0.3),
tf.keras.layers.Dense(units=1, activation='sigmoid')
])
model.load_weights('./checkpoint2.x/Titanic.100.h5')
model.compile(optimizer=tf.keras.optimizers.Adam(0.003),
loss='binary_crossentropy',
metrics=['accuracy'])
loss, acc = model.evaluate(x_test, y_test, verbose=2)
print('Restore model accuracy:{:5.4f}%'.format(100 * acc))
262/1 - 0s - loss: 0.5042 - accuracy: 0.8511
Restore model accuracy:85.1145%
Titanic TensorFLow 1.x Keras API 实现
导入必要的包
import numpy
import pandas as pd
import tensorflow as tf
import urllib.request
from sklearn import preprocessing
import matplotlib.pyplot as plt
import os
import datetime
tf.__version__
'1.15.2'
定义预处理函数
def prepare_data(df_data):
df = df_data.drop(['name'], axis=1)
age_mean = df['age'].mean()
df['age'] = df['age'].fillna(age_mean)
fare_mean = df['fare'].mean()
df['fare'] = df['fare'].fillna(fare_mean)
df['sex'] = df['sex'].map({'female':0, 'male':1}).astype(int)
df['embarked'] = df['embarked'].fillna('S')
df['embarked'] = df['embarked'].map({'C':0, 'Q':1, 'S':2}).astype(int)
ndarray_data = df.values
features = ndarray_data[:, 1:]
label = ndarray_data[:, 0]
minmax_scale = preprocessing.MinMaxScaler(feature_range=(0, 1))
norm_features = minmax_scale.fit_transform(features)
return norm_features, label
读取数据,制作数据集
dataframe = pd.read_excel('./data/titanic3.xls')
selected_cols= ['survived', 'name', 'pclass', 'sex', 'age', 'sibsp', 'parch', 'fare', 'embarked']
selected_dataframe = dataframe[selected_cols].copy()
selected_dataframe = selected_dataframe.sample(frac=1)
x_data, y_data = prepare_data(selected_dataframe)
train_size = int(len(x_data) * 0.8)
x_train = x_data[:train_size]
y_train = y_data[:train_size]
x_test = x_data[train_size:]
y_test = y_data[train_size:]
搭建模型
model = tf.keras.models.Sequential([
tf.keras.layers.Dense(units=64,
input_dim=7,
use_bias=True,
kernel_initializer='uniform',
bias_initializer='zeros',
activation='relu'),
tf.keras.layers.Dropout(rate=0.3),
tf.keras.layers.Dense(units=32, activation='sigmoid'),
tf.keras.layers.Dropout(rate=0.3),
tf.keras.layers.Dense(units=1, activation='sigmoid')
])
WARNING:tensorflow:From e:\anaconda3\envs\tensorflow1.x\lib\site-packages\tensorflow_core\python\keras\initializers.py:119: calling RandomUniform.__init__ (from tensorflow.python.ops.init_ops) with dtype is deprecated and will be removed in a future version.
Instructions for updating:
Call initializer instance with the dtype argument instead of passing it to the constructor
WARNING:tensorflow:From e:\anaconda3\envs\tensorflow1.x\lib\site-packages\tensorflow_core\python\ops\resource_variable_ops.py:1630: calling BaseResourceVariable.__init__ (from tensorflow.python.ops.resource_variable_ops) with constraint is deprecated and will be removed in a future version.
Instructions for updating:
If using Keras pass *_constraint arguments to layers.
model.summary()
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense (Dense) (None, 64) 512
_________________________________________________________________
dropout (Dropout) (None, 64) 0
_________________________________________________________________
dense_1 (Dense) (None, 32) 2080
_________________________________________________________________
dropout_1 (Dropout) (None, 32) 0
_________________________________________________________________
dense_2 (Dense) (None, 1) 33
=================================================================
Total params: 2,625
Trainable params: 2,625
Non-trainable params: 0
_________________________________________________________________
model.compile(optimizer=tf.keras.optimizers.Adam(0.003),
loss='binary_crossentropy',
metrics=['accuracy'])
WARNING:tensorflow:From e:\anaconda3\envs\tensorflow1.x\lib\site-packages\tensorflow_core\python\ops\nn_impl.py:183: where (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.
Instructions for updating:
Use tf.where in 2.0, which has the same broadcast rule as np.where
log_dir = os.path.join(
'logs1.x',
'train',
'plugins',
'profile',
datetime.datetime.now().strftime('%Y-%m-%d_%H-%M-%S'))
checkpoint_path = './checkpoint1.x/Titanic_{epoch:02d}-{val_loss:.2f}.ckpt'
callbacks = [tf.keras.callbacks.TensorBoard(log_dir=log_dir,
histogram_freq=2),
tf.keras.callbacks.ModelCheckpoint(filepath=checkpoint_path,
save_weights_only=True,
verbose=1,
period=5)]
WARNING:tensorflow:`period` argument is deprecated. Please use `save_freq` to specify the frequency in number of samples seen.
开始训练
train_history = model.fit(x=x_train,
y=y_train,
validation_split=0.2,
epochs=100,
batch_size=40,
callbacks=callbacks,
verbose=2)
部分训练如下
Train on 837 samples, validate on 210 samples
837/837 - 0s - loss: 0.4398 - acc: 0.8124 - val_loss: 0.4671 - val_acc: 0.7857
Epoch 80/100
Epoch 00080: saving model to ./checkpoint1.x/Titanic_80-0.47.ckpt
837/837 - 0s - loss: 0.4360 - acc: 0.8076 - val_loss: 0.4673 - val_acc: 0.7857
Epoch 81/100
837/837 - 0s - loss: 0.4307 - acc: 0.8005 - val_loss: 0.4703 - val_acc: 0.7905
Epoch 82/100
837/837 - 0s - loss: 0.4401 - acc: 0.7981 - val_loss: 0.4666 - val_acc: 0.8000
Epoch 83/100
837/837 - 0s - loss: 0.4311 - acc: 0.8017 - val_loss: 0.4678 - val_acc: 0.7952
Epoch 84/100
837/837 - 0s - loss: 0.4296 - acc: 0.8172 - val_loss: 0.4673 - val_acc: 0.8000
Epoch 85/100
Epoch 00085: saving model to ./checkpoint1.x/Titanic_85-0.46.ckpt
837/837 - 0s - loss: 0.4384 - acc: 0.8029 - val_loss: 0.4634 - val_acc: 0.7857
Epoch 86/100
837/837 - 0s - loss: 0.4345 - acc: 0.8076 - val_loss: 0.4666 - val_acc: 0.7905
Epoch 87/100
837/837 - 0s - loss: 0.4307 - acc: 0.8053 - val_loss: 0.4650 - val_acc: 0.8000
Epoch 88/100
837/837 - 0s - loss: 0.4394 - acc: 0.8148 - val_loss: 0.4638 - val_acc: 0.8000
Epoch 89/100
837/837 - 0s - loss: 0.4355 - acc: 0.8053 - val_loss: 0.4648 - val_acc: 0.8000
Epoch 90/100
Epoch 00090: saving model to ./checkpoint1.x/Titanic_90-0.46.ckpt
837/837 - 0s - loss: 0.4326 - acc: 0.8100 - val_loss: 0.4623 - val_acc: 0.8000
Epoch 91/100
837/837 - 0s - loss: 0.4387 - acc: 0.8029 - val_loss: 0.4658 - val_acc: 0.7905
Epoch 92/100
837/837 - 0s - loss: 0.4285 - acc: 0.8065 - val_loss: 0.4613 - val_acc: 0.7905
Epoch 93/100
837/837 - 0s - loss: 0.4355 - acc: 0.8088 - val_loss: 0.4656 - val_acc: 0.7905
Epoch 94/100
837/837 - 0s - loss: 0.4318 - acc: 0.8136 - val_loss: 0.4629 - val_acc: 0.7952
Epoch 95/100
Epoch 00095: saving model to ./checkpoint1.x/Titanic_95-0.46.ckpt
837/837 - 0s - loss: 0.4386 - acc: 0.7981 - val_loss: 0.4639 - val_acc: 0.8000
Epoch 96/100
837/837 - 0s - loss: 0.4346 - acc: 0.8041 - val_loss: 0.4647 - val_acc: 0.7857
Epoch 97/100
837/837 - 0s - loss: 0.4256 - acc: 0.8160 - val_loss: 0.4608 - val_acc: 0.8048
Epoch 98/100
837/837 - 0s - loss: 0.4357 - acc: 0.8029 - val_loss: 0.4613 - val_acc: 0.8000
Epoch 99/100
837/837 - 0s - loss: 0.4265 - acc: 0.8041 - val_loss: 0.4614 - val_acc: 0.7952
Epoch 100/100
Epoch 00100: saving model to ./checkpoint1.x/Titanic_100-0.46.ckpt
837/837 - 0s - loss: 0.4243 - acc: 0.8148 - val_loss: 0.4611 - val_acc: 0.8000
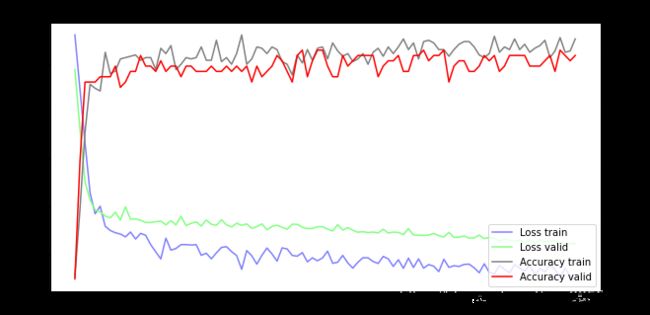
训练可视化
fig = plt.gcf()
fig.set_size_inches(10, 5)
ax1 = fig.add_subplot(111)
ax1.set_title('Train and Validation Picture')
ax1.set_ylabel('Loss value')
line1, = ax1.plot(train_history.history['loss'], color=(0.5, 0.5, 1.0), label='Loss train')
line2, = ax1.plot(train_history.history['val_loss'], color=(0.5, 1.0, 0.5), label='Loss valid')
ax2 = ax1.twinx()
ax2.set_ylabel('Accuracy value')
line3, = ax2.plot(train_history.history['acc'], color=(0.5, 0.5, 0.5), label='Accuracy train')
line4, = ax2.plot(train_history.history['val_acc'], color=(1, 0, 0), label='Accuracy valid')
plt.legend(handles=(line1, line2, line3, line4), loc='best')
plt.show()
模型预测
Jack_info = [0, 'Jack', 3, 'male', 23, 1, 0, 5.0000, 'S']
Rose_info = [1, 'Rose', 1, 'female', 20, 1, 0, 100.0000, 'S']
new_passenger_pd = pd.DataFrame([Jack_info, Rose_info], columns=selected_cols)
all_passenger_pd = selected_dataframe.append(new_passenger_pd)
pred = model.predict(prepare_data(all_passenger_pd)[0])
print('Rose survived probability:', pred[-1:][0][0],
'\nJack survived probability:', pred[-2:][0][0])
Rose survived probability: 0.9762004
Jack survived probability: 0.10789904
加载模型进行预测
model = tf.keras.models.Sequential([
tf.keras.layers.Dense(units=64,
input_dim=7,
use_bias=True,
kernel_initializer='uniform',
bias_initializer='zeros',
activation='relu'),
tf.keras.layers.Dropout(rate=0.3),
tf.keras.layers.Dense(units=32, activation='sigmoid'),
tf.keras.layers.Dropout(rate=0.3),
tf.keras.layers.Dense(units=1, activation='sigmoid')
])
model.compile(optimizer=tf.keras.optimizers.Adam(0.003),
loss='binary_crossentropy',
metrics=['accuracy'])
checkpoint_dir = os.path.dirname(checkpoint_path)
latest = tf.train.latest_checkpoint(checkpoint_dir)
model.load_weights(latest)
loss, acc = model.evaluate(x_test, y_test)
262/262 [==============================] - 0s 244us/sample - loss: 0.4393 - acc: 0.7977