Covid19:传染病模型模拟Agent based simulation for SIR/SEIR models
Content
- Scale-free Network Simulation
- Simulation for SIR model
- Codes for SIR simulation
- Simulation for SIR model
- Codes for SEIR simulation
- Reference
github地址
Scale-free Network Simulation
Simulation for SIR model
We conduct network simulation using Netlogo and construct a scale-free network using Netlogo extension nw.Here, we use a simulation with 2000 agents as an example.
We conduct an experiment to explore the range of the outcome if we put the parameters we pop in to a range. This is called parameter sensitivity test.
Here, we test the following parameters in the form of [minimum, step, maximum] in the model which run 500 times with 2000 agents and check the tick every 50 ticks.
| parameters | ranges |
|---|---|
| initial-outbreak-size | [1 1 5] |
| recovery-rate | [1 10 100] |
| virus-spread-rate | [1 10 100] |
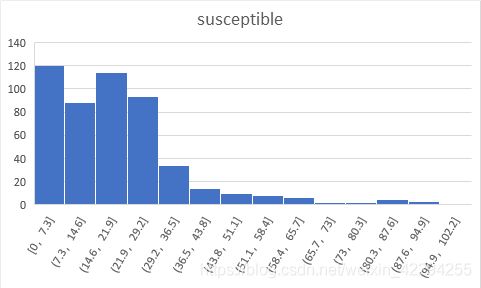
By running the model multiple times, we could get the average result of 500 time series data and calculate the minimum, maximum and mean results of the ratio of susceptible people, the ratio of infected people, the ratio of recovered people.
The simulation result show that the ratio of the three types of people follows power law distruibution which is exatly the property of the scale-free network.
| average(%) | min | max | mean |
|---|---|---|---|
| susceptible | 9.9720 | 97.0000 | 19.7972 |
| infected | 1.5560 | 78.2940 | 44.6495 |
| recovered | 0.0000 | 53.4540 | 35.5533 |
| virus | 1.5560 | 78.2940 | 44.6495 |
 |
 |
|---|
Codes for SIR simulation
extensions [ nw ]
;;; Extended model of AVI model for social propagation issues
;;; Concepts: Agent, virus, interaction, role, environment, social network
;;;
turtles-own ;; properties of agents;;
[
infected? ;; if true, the agent is infectious, role feature
resistant? ;; if true, the agent can't be infected, role feature
virus-recovered-period ;; number of ticks since this agent's last virus check, i.e., the minimum recovered period of agents
virus? ;; whether the agent carry the H1N1 virus
]
to setup
clear-all
setup-agents ;; setup of agents
setup-environment ;; setup of environment
degree-distribution-plot ;; chart plot setup
virus-status-plot ;; chart plot setup
virus-initialization
reset-ticks
end
to virus-initialization
ask n-of initial-outbreak-size turtles
[ play-infected ]
end
to setup-environment ;; environment setup, comprised of grids.
ask patches
[ set pcolor white ]
end
to setup-agents ;; agents setup;;
set-default-shape turtles "person"
nw:generate-preferential-attachment turtles links 100 [
;; Make agents not too close to each others.
setxy (random-xcor * 0.95) (random-ycor * 0.95)
set color green
play-susceptible
set virus-recovered-period random virus-check-frequency
]
end
to go
if all? turtles [not infected?]
[ stop ]
ask turtles
[
set virus-recovered-period virus-recovered-period + 1
if virus-recovered-period >= virus-check-frequency
[ set virus-recovered-period 0 ]
]
tick
interaction-between-agents
do-virus-checks
degree-distribution-plot
virus-status-plot
end
to interaction-between-agents ;; Interaction;interactions between agents, especially between the infected agents and the susceptiple agents
ask turtles with [infected?]
[
if count my-links > 0
[
ask one-of link-neighbors
[
;; interaction
if not resistant?
[ play-infected ]
]
]
]
end
to do-virus-checks
ask turtles with [infected? and virus-recovered-period = 0]
[
if random 100 < recovery-rate
[
ifelse random 100 < recovery-rate
[play-resistent]
[play-susceptible]
]
]
end
to play-infected ;; agent plays the infected role
set infected? true
set resistant? false
set virus? true
set color red
end
to play-susceptible ;; agent plays the susceptible role
set infected? false
set resistant? false
set virus? false
set color green
end
to play-resistent ;; agent plays the recovered role
set infected? false
set resistant? true
set virus? false
set color gray
ask my-links [ set color gray - 2 ]
end
to degree-distribution-plot
set-current-plot "Degree-distribution"
set-current-plot-pen "agent-percentage"
clear-plot
let max-degree 1
ask max-one-of turtles [count my-links]
[
set max-degree count my-links
]
let current_plot_degree 0
repeat max-degree + 1
[
plotxy current_plot_degree ( count turtles with [ (count my-links) = current_plot_degree ]/ (count turtles) ) * 100
set current_plot_degree current_plot_degree + 1
]
end
to virus-status-plot ;;; the chart for the status of H1N1 virus spread in the artificial society
set-current-plot "covid19-spread Status"
set-current-plot-pen "susceptible"
plot (count turtles with [not infected? and not resistant? ]) / (count turtles) * 100
set-current-plot-pen "infected"
plot (count turtles with [infected?]) / (count turtles) * 100
set-current-plot-pen "resistant"
plot (count turtles with [resistant?]) / (count turtles) * 100
;set-current-plot-pen "covid19 virus" ;;; This shows how many agents carry H1N1 virus
;plot (count turtles with [virus?]) / (count turtles) * 100
end
Simulation for SIR model
We conduct an experiment to explore the range of the outcome if we put the parameters we pop in to a range. This is called parameter sensitivity test.
Here, we test the following parameters in the form of [minimum, step, maximum] in the model which run 500 times with 2000 agents and check the tick every 50 ticks.
| parameters | ranges |
|---|---|
| initial-outbreak-size | [1 1 5] |
| recovery-rate | [1 10 100] |
| incubation-rate | [1 10 100] |
| virus-spread-rate | [1 10 100] |
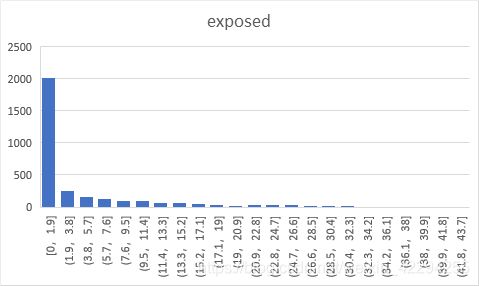
By running the model multiple times, we could get the average result of 500 time series data and calculate the minimum, maximum and mean results of the ratio of susceptible people, the ratio of exposed people, the ratio of infected people, the ratio of recovered people, and the ratio of the virus remaining in the system.
The simulation result show that the ratio of the four types of people follows power law distruibution which is exatly the property of the scale-free network.
| average(%) | min | max | mean |
|---|---|---|---|
| susceptible | 32.3918 | 97.8309 | 43.9674 |
| exposed | 0.0000 | 14.3373 | 5.7009 |
| infected | 1.2026 | 41.5529 | 21.8660 |
| recovered | 0.0000 | 46.8978 | 28.4640 |
| virus | 1.3938 | 48.6874 | 27.5569 |
 |
 |
|---|---|
 |
 |
Codes for SEIR simulation
extensions [ nw ]
;;; Extended model of AVI model for social propagation issues
;;; Concepts: Agent, virus, interaction, role, environment, social network
;;;
turtles-own ;; properties of agents;;
[
infected? ;; if true, the agent is infectious, role feature
resistant? ;; if true, the agent can't be infected, role feature
exposed? ;; if true, the agent is exposed, role feature
recovered? ;; if true, the agent is recovered, role feature
virus-recovered-period ;; number of ticks since this agent's last virus check, i.e., the minimum recovered period of agents
virus? ;; whether the agent carry the H1N1 virus
]
to setup
__clear-all-and-reset-ticks
setup-agents ;; setup of agents
setup-environment ;; setup of environment
degree-distribution-plot ;; chart plot setup
virus-status-plot ;; chart plot setup
virus-initialization
end
to virus-initialization
ask n-of initial-outbreak-size turtles
[ play-infected ]
end
to setup-environment ;; environment setup, comprised of grids.
ask patches
[ set pcolor white ]
end
to setup-agents ;; agents setup;;
set-default-shape turtles "person"
nw:generate-preferential-attachment turtles links 100 [
;; Make agents not too close to each others.
setxy (random-xcor * 0.95) (random-ycor * 0.95)
set color green
play-susceptible
set virus-recovered-period random virus-check-frequency
]
end
;how does the JGN social network danymically grow?
;construct the network
to go
if ( ticks > simulation-num )
[stop]
ask turtles
[
set virus-recovered-period virus-recovered-period + 1
if virus-recovered-period >= virus-check-frequency
[ set virus-recovered-period 0 ]
]
interaction-between-agents
tick
do-virus-checks
degree-distribution-plot
virus-status-plot
end
to interaction-between-agents ;; Interaction;interactions between agents, especially between the infected agents and the susceptiple agents
ask turtles with [infected?]
[
if count my-links > 0
[
ask one-of link-neighbors
[
;; interaction
if not recovered? and random-float 100 < incubation-rate
[ play-exposed ]
]
]
]
end
to do-virus-checks
ask turtles with [infected? and virus-recovered-period = 0]
[
if random 100 < recovery-rate
[
play-recovered
]
]
ask turtles with [exposed?]
[
if random-float 100 < virus-spread-rate
[play-infected]
]
end
to play-exposed ;; agent plays the exposed role
set infected? false
set exposed? true
set recovered? false
set virus? true
set virus-recovered-period random virus-check-frequency
set color blue
end
to play-infected ;; agent plays the infected role
set infected? true
set exposed? false
set recovered? false
set virus? true
set color red
end
to play-susceptible ;; agent plays the susceptible role
set infected? false
set exposed? false
set recovered? false
set virus? false
set color green
end
to play-recovered ;; agent plays the recovered role
set infected? false
set exposed? false
set recovered? true
set virus? false
set color gray
ask my-links [ set color gray - 2 ]
end
to degree-distribution-plot
set-current-plot "Degree-distribution"
set-current-plot-pen "agent-percentage"
clear-plot
let max-degree 1
ask max-one-of turtles [count my-links]
[
set max-degree count my-links
]
let current_plot_degree 0
repeat max-degree + 1
[
plotxy current_plot_degree ( count turtles with [ (count my-links) = current_plot_degree ]/ (count turtles) ) * 100
set current_plot_degree current_plot_degree + 1
]
end
to virus-status-plot ;;; the chart for the status of H1N1 virus spread in the artificial society
set-current-plot "covid19-spread Status"
set-current-plot-pen "susceptible"
plot (count turtles with [not infected? and not recovered? and not exposed?]) / (count turtles) * 100
set-current-plot-pen "infected"
plot (count turtles with [infected?]) / (count turtles) * 100
set-current-plot-pen "recovered"
plot (count turtles with [recovered?]) / (count turtles) * 100
set-current-plot-pen "exposed"
plot (count turtles with [exposed?])/(count turtles) * 100
; set-current-plot-pen "covid19 virus" ;;; This shows how many agents carry H1N1 virus
;plot (count turtles with [virus?]) / (count turtles) * 100
end
Reference
.
Stonedahl, F. and Wilensky, U. (2008). NetLogo Virus on a Network model.
Center for Connected Learning and Computer-Based Modeling, Northwestern University, Evanston, IL.Wilensky, U. (1999). NetLogo.
Center for Connected Learning and Computer-Based Modeling, Northwestern University, Evanston, IL.
欢迎关注二幺子的知识输出通道:


