前言
昨天CJ微信推送pfam的本地安装,以下为原文链接:
pfam安装和使用:https://www.jianshu.com/p/9cf40d0d8bf5
目前我做lncRNA分析,在线使用不太方便,刚准备搭建pfam本地安装,嘻嘻~
原文已经很详细,但是第一次使用,我一些疑问,记录了自己的安装过程,所以有了下面的内容
Pfam是一个大型蛋白结构域家族的数据库,每个蛋白家族都由多个序列比对和HMMs(hidden Markovmodels,隐马尔可夫模型)所体现。Pfam包括两个质量级别的家族数据库:Pfam-A和Pfam-B(pfam27.0包含,以后更新的版本没有)。Pfam-A来自基础序列数据库Pfamseq,是根据最新的UniProtKB数据建立的,质量较高。Pfam-B做为Pfam-A的补充,是一个未注释的低质量数据库,一般是由ADDA数据中的非冗余cluster自动生成的。虽然质量较低,但对于鉴定Pfam-A无法覆盖到的功能保守区域是非常有用的。
pfam http://ftp.ebi.ac.uk/pub/databases/Pfam/releases/
1.PfamScan本地安装:Tools文件夹(我下载的最新版)
1)解压,不需要修改开头指定perl
pengzw@super-server:~$ mkdir ~/biosoft/pfam
pengzw@super-server:~$ cd ~/biosoft/pfam
pengzw@super-server:~/biosoft/pfam$ nohup wget ftp://ftp.ebi.ac.uk/pub/databases/Pfam/releases/Pfam32.0/Pfam-A.hmm.gz &
pengzw@super-server:~/biosoft/pfam$ nohup wget http://ftp.ebi.ac.uk/pub/databases/Pfam/Tools/PfamScan.tar.gz &
pengzw@super-server:~/biosoft/pfam$ tar zxvf PfamScan.tar.gz
pengzw@super-server:~/biosoft/pfam$ cd PfamScan
pengzw@super-server:~/biosoft/pfam$ cat
pengzw@super-server:~/biosoft/pfam/PfamScan$ cat pfam_scan.pl |head -n 5
#!/usr/bin/env perl
# $Id: pfam_scan.pl 9592 2017-02-23 09:32:06Z jaina $
把Pfam目录加入环境,pfam_scan.pl所在目录
在末尾加入export PERL5LIB=/home/pengzw/biosoft/pfam/PfamScan/:$PERL5LIB
pengzw@super-server:~/biosoft/pfam/PfamScan$ vim ~/.bashrc
2.安装hmmer3 (之前安装过,v3.2.1)
3.下载Pfam库(我选的最新版pfam32.0),放在同一目录PfamData,否则会出错!!!
安装
pengzw@super-server:~/biosoft/pfam$ mkdir PfamData
pengzw@super-server:~/biosoft/pfam$ cd PfamData
pengzw@super-server:~/biosoft/pfam/PfamData$ nohup wget ftp://ftp.ebi.ac.uk/pub/databases/Pfam/releases/Pfam32.0/Pfam-A.hmm.dat.gz &
pengzw@super-server:~/biosoft/pfam/PfamData$ nohup wget ftp://ftp.ebi.ac.uk/pub/databases/Pfam/Tools/PfamScan.tar.gz &
pengzw@super-server:~/biosoft/pfam/PfamData$ gunzip Pfam-A.hmm.gz Pfam-A.hmm.dat.gz
pengzw@super-server:~/biosoft/pfam/PfamData$ hmmpress Pfam-A.hmm
使用
pengzw@super-server:~/biosoft/pfam$ ./PfamScan/pfam_scan.pl -h
pfam_scan.pl: search a FASTA file against a library of Pfam HMMs
Usage: pfam_scan.pl -fasta -dir
Additonal options:
-h : show this help
-outfile : output file, otherwise send to STDOUT
-clan_overlap : show overlapping hits within clan member families (applies to Pfam-A families only)
-align : show the HMM-sequence alignment for each match
-e_seq : specify hmmscan evalue sequence cutoff for Pfam-A searches (default Pfam defined)
-e_dom : specify hmmscan evalue domain cutoff for Pfam-A searches (default Pfam defined)
-b_seq : specify hmmscan bit score sequence cutoff for Pfam-A searches (default Pfam defined)
-b_dom : specify hmmscan bit score domain cutoff for Pfam-A searches (default Pfam defined)
-as : predict active site residues for Pfam-A matches
-json [pretty] : write results in JSON format. If the optional value "pretty" is given,
the JSON output will be formatted using the "pretty" option in the JSON
module
-cpu : number of parallel CPU workers to use for multithreads (default all)
-translate [mode] : treat sequence as DNA and perform six-frame translation before searching. If the
optional value "mode" is given it must be either "all", to translate everything
and produce no individual ORFs, or "orf", to report only ORFs with length greater
than 20. If "-translate" is used without a "mode" value, the default is to
report ORFs (default no translation)
pengzw@super-server:~/biosoft/pfam$ ./PfamScan/pfam_scan.pl -fasta AtCBL.fa -dir ./PfamData/ -out test.fa
pengzw@super-server:~/biosoft/pfam$ cat test2.fa
# pfam_scan.pl, run at Wed Apr 3 11:28:40 2019
#
# Copyright (c) 2009 Genome Research Ltd
# Freely distributed under the GNU
# General Public License
#
# Authors: Jaina Mistry ([email protected]),
# Rob Finn ([email protected])
#
# This is free software; you can redistribute it and/or modify it under
# the terms of the GNU General Public License as published by the Free Software
# Foundation; either version 2 of the License, or (at your option) any later version.
# This program is distributed in the hope that it will be useful, but WITHOUT
# ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS
# FOR A PARTICULAR PURPOSE. See the GNU General Public License for more
# details.
#
# You should have received a copy of the GNU General Public License along with
# this program. If not, see
AtCBL8 53 98 52 102 PF13833.6 EF-hand_8 Domain 2 50 55 14.1 0.031 1 CL0220
AtCBL8 115 179 111 182 PF13499.6 EF-hand_7 Domain 6 68 71 38.7 1e-09 1 CL0220
AtCBL5 45 97 26 100 PF12763.7 EF-hand_4 Family 20 71 104 22.3 9.2e-05 1 CL0220
AtCBL5 107 175 105 177 PF13499.6 EF-hand_7 Domain 3 69 71 30.3 4.2e-07 1 CL0220
AtCBL6 116 181 115 187 PF13499.6 EF-hand_7 Domain 2 65 71 36.1 6.6e-09 1 CL0220
AtCBL1 107 172 106 179 PF13499.6 EF-hand_7 Domain 2 65 71 31.7 1.5e-07 1 CL0220
AtCBL7 77 100 75 103 PF13405.6 EF-hand_6 Domain 4 27 31 23.4 3.6e-05 1 CL0220
AtCBL7 156 180 155 180 PF13202.6 EF-hand_5 Domain 1 25 25 15.1 0.011 1 CL0220
AtCBL3 64 113 63 116 PF13833.6 EF-hand_8 Domain 2 50 55 21.0 0.00021 1 CL0220
AtCBL3 126 193 125 197 PF13499.6 EF-hand_7 Domain 2 67 71 34.9 1.5e-08 1 CL0220
AtCBL10 119 138 118 143 PF13202.6 EF-hand_5 Domain 2 21 25 13.6 0.034 1 CL0220
AtCBL10 153 218 152 223 PF13499.6 EF-hand_7 Domain 2 65 71 38.5 1.2e-09 1 CL0220
AtCBL4 111 180 110 182 PF13499.6 EF-hand_7 Domain 2 69 71 35.0 1.5e-08 1 CL0220
AtCBL9 153 177 153 177 PF13202.6 EF-hand_5 Domain 1 25 25 28.5 6.7e-07 1 CL0220
AtCBL2 64 109 63 112 PF13833.6 EF-hand_8 Domain 2 50 55 21.5 0.00015 1 CL0220
AtCBL2 122 187 121 192 PF13499.6 EF-hand_7 Domain 2 65 71 36.1 6.8e-09 1 CL0220
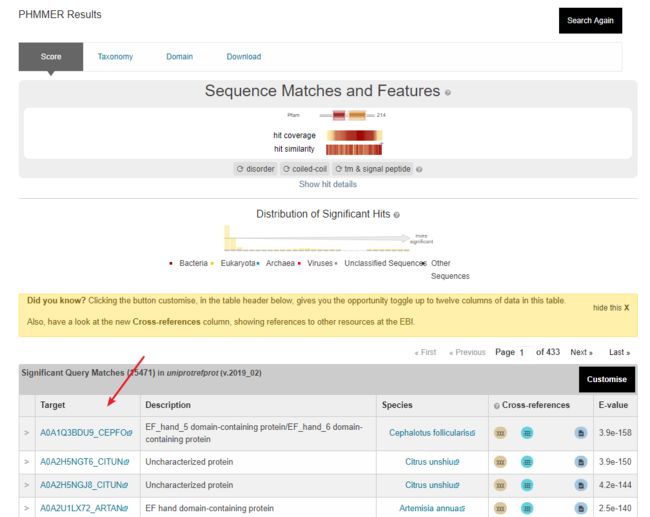
4.在线:pfam search在线结果,邮箱查收(部分浏览器可以,我用的google)~
1.网页结果,直接调到HMMER界面
2.邮箱结果
5.ps最后:原来我用了这么久的HMMSCAN,表示由于某些原因受到了欺骗,还是要多思考。。。我还一直以为HMMER有个在线数据库~
phmmer和hmmscan的结果差别如下,其实我还是用的pfam的结果啊,只是利用了hmmer这个软件。
6.参考原文:
pfam安装和使用:https://www.jianshu.com/p/9cf40d0d8bf5
Pfam数据库蛋白编码能力预测http://www.biotrainee.com/thread-1212-1-1.html