FSL、MRICRON预处理DICOM数据,python读取nii,并降采样
1.FSL和MRICRON预处理DICOM数据
软件安装见:https://blog.csdn.net/qq_29477893/article/details/89376758。推荐第二种手动安装方法,尝试过几次都一次性安装成功。
步骤:
1)dicom数据转换为nii数据
2)FSL的bet提取大脑
3)FSL的flirt配准到标准大脑模板(/usr/local/fsl/data/standard/MNI152_T1_1mm_brain.nii.gz)
批处理shell脚本
#!/bin/bash
ref="/usr/local/fsl/data/standard/MNI152_T1_1mm_brain.nii.gz" # 标准化空间模板
#mask_ref="/PUBLIC/wangcong/software/model/cerebellar_mask.nii" #标准小脑mask模板
precessing(){
for subject in $(ls $1)
do
echo "处理$1/..."
echo $subject
start_time=`date +%s`
#准备预处理文件夹
input_mri=$1/$subject
temp_mri=$2/$subject
output_mri=$3
mkdir -p $temp_mri
mkdir -p $output_mri
#开始预处理步骤
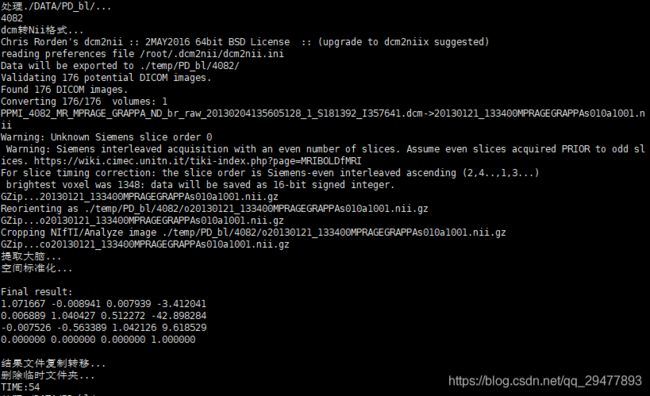
echo "dcm转Nii格式..."
dcm2nii -o $temp_mri $input_mri #dcm转Nii格式,输出到temp_dti
raw_mri=`ls $temp_mri/co*.nii.gz` #找到需要的文件名
echo "提取大脑..."
brain_mri="brain_mri" #输出大脑文件的命名
bet $raw_mri $temp_mri/$brain_mri -R #提取大脑,参数-R
echo "空间标准化..."
norm_mri="norm_mri"
flirt -in $temp_mri/${brain_mri}.nii.gz -ref $ref -out $temp_mri/$norm_mri #模板文件ref
echo "结果文件复制转移..."
out_mri=${subject}_mri
cp $temp_mri/${norm_mri}.nii.gz $output_mri/${out_mri}.nii.gz
echo "删除临时文件夹..."
rm -rf $2/$subject
stop_time=`date +%s`
echo "TIME:`expr $stop_time - $start_time`"
done
}
input="./DATA/$1" #原始文件夹
temp="./temp/$1" #临时文件夹
output="./processed/$1" #结果文件夹
precessing $input $temp $output
2.使用python的nibabel库显示处理图像
下面代码包含了三个函数,读取函数,显示函数,降采样函数
# -*- coding: utf-8 -*-
"""
Created on Thu Jul 2 10:16:18 2020
@author: cong
"""
import nibabel as nib
import matplotlib.pyplot as plt
import numpy as np
def read_data(path):
image_data = nib.load(path).get_data()
return image_data
def show_img(ori_img,c):
if c==1:
plt.imshow(ori_img[:,:,ori_img.shape[2]//2], cmap = 'gray') #channel_last
plt.show()
if c==2:
plt.imshow(ori_img[:,ori_img.shape[1]//2,:], cmap = 'gray') #channel_last
plt.show()
if c==3:
plt.imshow(ori_img[ori_img.shape[0]//2,:,:], cmap = 'gray') #channel_last
plt.show()
def down_sample(data,n1,n2,n3):#n1,n2,n3各个维度的降采样比例
[r,c,h] = data.shape
dr = round(r/n1)
dc = round(c/n2)
dh = round(h/n3)
Idown = np.zeros((dr,dc,dh))
p =0
q =0
w =0
for i in range(0,r,n1):
for j in range(0,c,n2):
for k in range(0,h,n3):
Idown[p,q,w] = data[i,j,k]
w = w+1
w =0
q = q+1
q = 0
p = p+1
return Idown
#读取并显示原始图像
path = '../processed/PD_bl/3102_mri.nii.gz'
data = read_data(path)
print(data.shape)
show_img(data,1)
show_img(data,2)
show_img(data,3)
#降采样图像并显示
down_data=down_sample(data,2,2,2)
print(down_data.shape)
show_img(down_data,1)
show_img(down_data,2)
show_img(down_data,3)
#保存处理后的array为nii数据
affine = np.diag([1, 2, 3, 1])
new_image = nib.Nifti1Image(down_data,affine)
nib.save(new_image, 'downsample_3102_mri.nii.gz')
#读取显示降采样后的图像(确认无误)
down_data = read_data('downsample_3102_mri.nii.gz')
show_img(down_data,1)
show_img(down_data,2)
show_img(down_data,3)