渐进式学习:如何用R和GO富集可视化捕捉生命的关键信号?
一、引言
生命科学中的数据分析和可视化是一个具有挑战性的领域。随着技术和理论的不断发展,研究人员需要处理越来越复杂和庞大的数据集,以研究生物体在不同尺度上的结构和功能,探索不同生物过程和疾病的机制。在这个领域,GO(Gene Ontology)富集分析已成为一种常见的技术,用于识别在给定的基因集合中与特定生物过程和机制有关的基因集合,是研究生物信息学、基因组学和转录组学的重要方法之一[1,2]。然而,GO富集分析的结果往往是一张庞大的表格,需要通过可视化才能更好地理解和分析。在这方面,R语言作为一个强大的数据分析工具,也是生命科学研究领域中广泛使用的计算统计工具之一[3,4]。因此,本文将介绍GO富集分析技术并重点介绍R语言及其绘图包,如ggplot2和clusterProfiler等,用于可视化GO富集分析结果[5,6,7]。本文还将提供一些使用R和GO富集可视化的基本方法和技巧,并以实例说明如何从生物大数据中捕捉关键信号。最后,我们将讨论GO富集可视化在生物信息学中的未来发展和可能的研究方向。
二、数据集
2.1 数据集导入
该数据集是GOplot自带的数据集,可以用来学习Go富集分析和可视化,接下来我们引入数据集
library(GOplot)
data(EC)
数据集展示:
> # 查看数据
> head(EC$eset)
Gene_Symbol Brain_A Brain_B Brain_C Heart_A Heart_B Heart_C
1 0610007P14Rik 0.58382130 0.81117820 -0.2480545 -0.67075443 -0.58700850 -0.65176487
2 0610008F07Rik 0.13262606 0.11230135 0.3122339 0.10193062 -0.06617737 0.09765196
3 0610009B22Rik -0.09357643 -0.20074654 0.2505083 -1.02136140 -0.18762684 -0.69150734
4 0610009D07Rik 0.06708336 0.12616920 -0.1234570 -0.74400520 -0.42762280 -0.43724203
5 0610009O20Rik 0.02191877 0.13460398 0.2718530 0.27460957 -0.07751560 0.00000000
6 0610010F05Rik -0.10588837 -0.05568028 0.0115509 0.06084633 -0.18752098 0.01155090
> # 查看数据
> head(EC$genelist)
ID logFC AveExpr t P.Value adj.P.Val B
1 Slco1a4 6.645388 1.2168670 88.65515 1.32e-18 2.73e-14 29.02715
2 Slc19a3 6.281525 1.1600468 69.95094 2.41e-17 2.49e-13 27.62917
3 Ddc 4.483338 0.8365231 65.57836 5.31e-17 3.65e-13 27.18476
4 Slco1c1 6.469384 1.3558865 59.87613 1.62e-16 8.34e-13 26.51242
5 Sema3c 5.515630 2.3252117 58.53141 2.14e-16 8.81e-13 26.33626
6 Slc38a3 4.761755 0.9218670 54.11559 5.58e-16 1.76e-12 25.70308
> # 查看数据
> head(EC$david)
Category ID Term
1 BP GO:0007507 heart development
2 BP GO:0001944 vasculature development
3 BP GO:0001568 blood vessel development
4 BP GO:0048729 tissue morphogenesis
5 BP GO:0048514 blood vessel morphogenesis
6 BP GO:0051336 regulation of hydrolase activity
Genes
1 DLC1, NRP2, NRP1, EDN1, PDLIM3, GJA1, TTN, GJA5, ZIC3, TGFB2, CERKL, GATA6, COL4A3BP, GAB1, SEMA3C, MKL2, SLC22A5, MB, PTPRJ, RXRA, VANGL2, MYH6, TNNT2, HHEX, MURC, MIB1, FOXC2, FOXC1, ADAM19, MYL2, TCAP, EGLN1, SOX9, ITGB1, CHD7, HEXIM1, PKD2, NFATC4, PCSK5, ACTC1, TGFBR2, NF1, HSPG2, SMAD3, TBX1, TNNI3, CSRP3, FOXP1, KCNJ8, PLN, TSC2, ATP6V0A1, TGFBR3, HDAC9
2 GNA13, ACVRL1, NRP1, PGF, IL18, LEPR, EDN1, GJA1, FOXO1, GJA5, TGFB2, WARS, CERKL, APOE, CXCR4, ANG, SEMA3C, NOS2, MKL2, FGF2, RAPGEF1, PTPRJ, RECK, EFNB2, VASH1, PNPLA6, THY1, MIB1, NUS1, FOXC2, FOXC1, CAV1, CDH2, MEIS1, WT1, CDH5, PTK2, FBXW8, CHD7, PLCD1, PLXND1, FIGF, PPAP2B, MAP2K1, TBX4, TGFBR2, NF1, TBX1, TNNI3, LAMA4, MEOX2, ECSCR, HBEGF, AMOT, TGFBR3, HDAC7
3 GNA13, ACVRL1, NRP1, PGF, IL18, LEPR, EDN1, GJA1, FOXO1, GJA5, TGFB2, WARS, CERKL, APOE, CXCR4, ANG, SEMA3C, NOS2, MKL2, FGF2, RAPGEF1, PTPRJ, RECK, VASH1, PNPLA6, THY1, MIB1, NUS1, FOXC2, FOXC1, CAV1, CDH2, MEIS1, WT1, CDH5, PTK2, FBXW8, CHD7, PLCD1, PLXND1, FIGF, PPAP2B, MAP2K1, TBX4, TGFBR2, NF1, TBX1, TNNI3, LAMA4, MEOX2, ECSCR, HBEGF, AMOT, TGFBR3, HDAC7
4 DLC1, ENAH, NRP1, PGF, ZIC2, TGFB2, CD44, ILK, SEMA3C, RET, AR, RXRA, VANGL2, LEF1, TNNT2, HHEX, MIB1, NCOA3, FOXC2, FOXC1, TGFB1I1, WNT5A, COBL, BBS4, FGFR3, TNC, BMPR2, CTNND1, EGLN1, NR3C1, SOX9, TCF7L1, IGF1R, FOXQ1, MACF1, HOXA5, BCL2, PLXND1, CAR2, ACTC1, TBX4, SMAD3, FZD3, SHANK3, FZD6, HOXB4, FREM2, TSC2, ZIC5, TGFBR3, APAF1
5 GNA13, CAV1, ACVRL1, NRP1, PGF, IL18, LEPR, EDN1, GJA1, CDH2, MEIS1, WT1, TGFB2, WARS, PTK2, CERKL, APOE, CXCR4, ANG, SEMA3C, PLCD1, NOS2, MKL2, PLXND1, FIGF, FGF2, PTPRJ, TGFBR2, TBX4, NF1, TBX1, TNNI3, PNPLA6, VASH1, THY1, NUS1, MEOX2, ECSCR, AMOT, HBEGF, FOXC2, FOXC1, HDAC7
6 CAV1, XIAP, AGFG1, ADORA2A, TNNC1, TBC1D9, LEPR, ABHD5, EDN1, ASAP2, ASAP3, SMAP1, TBC1D12, ANG, TBC1D14, MTCH1, TBC1D13, TBC1D4, TBC1D30, DHCR24, HIP1, VAV3, NOS1, NF1, MYH6, RICTOR, TBC1D22A, THY1, PLCE1, RNF7, NDEL1, CHML, IFT57, ACAP2, TSC2, ERN1, APAF1, ARAP3, ARAP2, ARAP1, HTR2A, F2R
adj_pval
1 0.000002170
2 0.000010400
3 0.000007620
4 0.000119000
5 0.000720000
6 0.001171166
> # 查看数据
> head(EC$genes)
ID logFC
1 PTK2 -0.6527904
2 GNA13 0.3711599
3 LEPR 2.6539788
4 APOE 0.8698346
5 CXCR4 -2.5647537
6 RECK 3.6926860
> # 查看数据
> EC$process
[1] "heart development" "phosphorylation" "vasculature development" "blood vessel development" "tissue morphogenesis" "cell adhesion"
[7] "plasma membrane"
2.2 数据预处理
使用cirlce_dat函数整合GO注释结果数据和基因差异表达分析数据结合起来,形成作图对象。
circ <- circle_dat(EC$david, EC$genelist)
head(circ)
结果展示:
category ID term count genes logFC adj_pval zscore
1 BP GO:0007507 heart development 54 DLC1 -0.9707875 2.17e-06 -0.8164966
2 BP GO:0007507 heart development 54 NRP2 -1.5153173 2.17e-06 -0.8164966
3 BP GO:0007507 heart development 54 NRP1 -1.1412315 2.17e-06 -0.8164966
4 BP GO:0007507 heart development 54 EDN1 1.3813006 2.17e-06 -0.8164966
5 BP GO:0007507 heart development 54 PDLIM3 -0.8876939 2.17e-06 -0.8164966
6 BP GO:0007507 heart development 54 GJA1 -0.8179480 2.17e-06 -0.8164966
「字段简单介绍」:
-
category:GO term分为3大类,分别为BP(生物学过程),CC(细胞组分)和MF(分子功能) - ID和term来属于数据库的字段;
- count为划分到相应term的基因数目;
- logFC为差异表达基因的log标准化的倍数变化; adj_pval为校正过的p-value值,adj_pval < 0.05显著富集;
三、数据可视化
3.1 柱形图
- 所有种类
GOBar(circ, display = 'multiple', zsc.col = c('blue', 'white', 'red'))
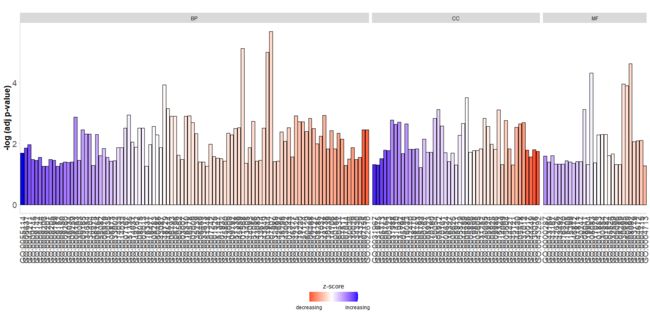
是不是觉得有写不清晰,看不出x坐标的标注。没关系,我们可以选择性展示其中感兴趣的部分。
- 展示特定种类
GOBar(subset(circ, category == 'MF'))
3.2 气泡图
GOBubble(circ, labels = 2.8)
 labels是一个很重要的参数,表示标记adj_pval负对数大于等于设置值的GO term,气泡的大小表明富集到该GO term的基因数目。我设置成了2.8,可读性还是可以的,但是交叉的气泡很多,我们可以通过
labels是一个很重要的参数,表示标记adj_pval负对数大于等于设置值的GO term,气泡的大小表明富集到该GO term的基因数目。我设置成了2.8,可读性还是可以的,但是交叉的气泡很多,我们可以通过reduce_overlap函数过滤一部分数据,减少泡泡的数目,如下:
reduced_circ <- reduce_overlap(circ, overlap = 0.70)
GOBubble(reduced_circ, labels = 2.8)
 视觉效果是不是比之前是要好一些!如果还是比较拥挤,我们可以分组绘图,如下:
视觉效果是不是比之前是要好一些!如果还是比较拥挤,我们可以分组绘图,如下:
GOBubble(circ, title = 'Bubble plot', display = 'multiple', labels = 3)

3.3 环形图
GOCircle(circ)

3.4 弦图
数据整理:
chord <- chord_dat(circ, EC$genes, EC$process)
head(chord)
结果展示:
> head(chord)
heart development phosphorylation vasculature development blood vessel development tissue morphogenesis cell adhesion plasma membrane logFC
PTK2 0 1 1 1 0 0 1 -0.6527904
GNA13 0 0 1 1 0 0 1 0.3711599
LEPR 0 0 1 1 0 0 1 2.6539788
APOE 0 0 1 1 0 0 1 0.8698346
CXCR4 0 0 1 1 0 0 1 -2.5647537
RECK 0 0 1 1 0 0 1 3.6926860
画图:
GOChord(chord, space = 0.02, gene.order = 'logFC', gene.space = 0.25, gene.size = 5)
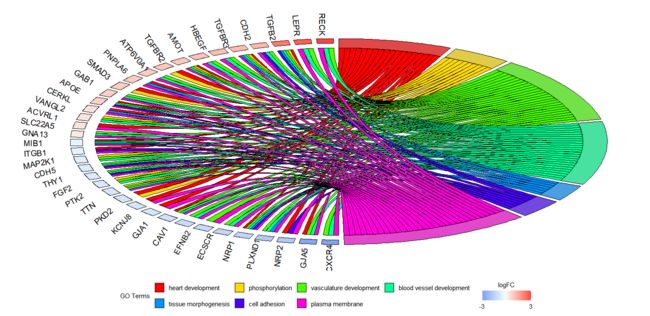 弦图展示了感兴趣的基因和某些GO term的关系,以基因差异表达倍数的大小排序。
弦图展示了感兴趣的基因和某些GO term的关系,以基因差异表达倍数的大小排序。
3.5 热图
GOHeat(chord, nlfc = 1, fill.col = c('red', 'blue', 'green'))

3.6 聚类图
GOCluster(circ, EC$process, clust.by = 'logFC', term.width = 2)

3.7 韦恩图
l1 <- subset(circ, term == 'heart development', c(genes,logFC))
l2 <- subset(circ, term == 'plasma membrane', c(genes,logFC))
l3 <- subset(circ, term == 'tissue morphogenesis', c(genes,logFC))
GOVenn(l1,l2,l3, label = c('heart development', 'plasma membrane', 'tissue morphogenesis'))

四、总结
本文介绍了如何使用R和GO富集可视化技术,分析生物信息学中数据集的复杂性问题。通过R语言和相应的包,如ggplot2,g:Profiler,moduleColor等,可以对生成的结果进行高效而准确的可视化,以帮助研究人员更好地理解和分析复杂数据集。文章还提供了一些使用R和GO富集可视化技术的基本方法和技巧,并以实例说明如何从生物大数据中捕捉关键信号。最后,我们探讨了GO富集可视化的未来趋势和展望,旨在为生命科学研究者提供更好的技术支持,以研究更有意义、有价值的数据。R语言在数据分析和可视化领域的应用将有望为大规模数据集的处理和分析,提供更有效、更快捷、更精确的解决方法。
参考文献:
[1] Ashburner M, Ball C A, Blake J A, et al. Gene ontology: tool for the unification of biology. Nature genetics, 2000, 25(1): 25-29.
[2] Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research, 2000, 28(1): 27-30.
[3] Gentleman R C, Carey V J, Bates D M, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology, 2004, 5(10): R80.
[4] R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria (2017). Available online at https://www.R-project.org/.
[5] Yu G, Wang L G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology, 2012, 16(5): 284-287.
[6] Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer, 2016.
[7] Kolde R, Laur S, Adler P, et al. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics, 2012, 28(4): 573-580.
