PSP - 蛋白质复合物结构预测 Template 的 Multichain Mask 2D (二维多链掩码)
欢迎关注我的CSDN:https://spike.blog.csdn.net/
本文地址:https://spike.blog.csdn.net/article/details/134406459
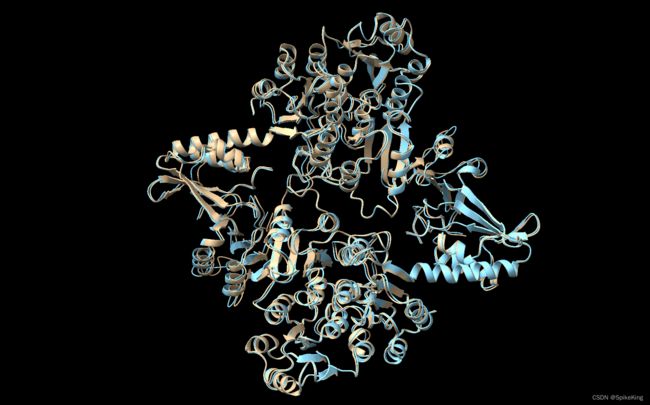
在 蛋白质复合物结构预测 中,AlphaFold2 Multimer 的 Multichain Mask 2D 对于 模版特征 (Template) 的影响较大,默认使用单链进行模版搜索,关闭链间 Docking 信息,只保留链内信息,当单链来源自同一个模版时,则可以保留链间信息。
测试 Case,8BBY:
>A
MSKVETGDQGYTVVQSKYKKAVEQLQKGLLDGEIKIFFEGTLASTIYCLHKVDNKLDNLGDGDYVDFLIITKLRILNAKEETIDIDASSSKTAQDLAKKYVFNKTDLNTLYRVLNGDEADTNRLVEEVSGKYQVVLYPEGKRV
>B
AAKASIADENSPVKLTLKSDKKKDLKDYVDDLRTYNNGYSNAIEVAGEDRIETAIALSQKYYNSDDENAIFRDSVDNVVLVGGNAIVDGLVASPLASEKKAPLLLTSKDKLDSSVKAEIKRVMNIKSTTGINTSKKVYLAGGVNSISKEVENELKDMGLKVTRLAGDDRYETSLKIADEVGLDNDKAFVVGGTGLADAMSIAPVASQLRNANGKMDLADGDATPIVVVDGKAKTINDDVKDFLDDSQVDIIGGENSVSKDVENAIDDATGKSPDRYSGDDRQATNAKVIKESSYYQDNLNNDKKVVNFFVAKDGSTKEDQLVDALAAAPVAANFGVTLNSDGKPVDKDGKVLTGSDNDKNKLVSPAPIVLATDSLSSDQSVSISKVLDKDNGENLVQVGKGIATSVINKLKDLLSM
>C
DMSKVETGDQGYTVVQSKYKKAVEQIKIFFEGTLAYCLHKVDNKLDNLGDGDYVDFLIITKLRILNAKEETIDIDASSSKTAQDLAKKYVFNKTDLNTLYRVLNGDEADTNRVEEVSGKYQVVLYPEGKRV
>D
ASIADENSPVKLTLKSDKKKDLKDYVDDLRTYNNGYSNAIEVAGEDRIETAIALSQKYYNSDDENAIFRDSVDNVVLVGGNAIVDGLVASPLASEKKAPLLLTSKDKLDSSVKAEIKRVMNIKSTTGINTSKKVYLAGGVNSISKEVENELKDMGLKVTRLAGDDRYETSLKIADEVGLDNDKAFVVGGTGLADAMSIAPVASQLRNANGKMDLADGDATPIVVVDGKAKTINDDVKDFLDDSQVDIIGGENSVSKDVENAIDDATGKSPDRYSGDDRQATNAKVIKESSYYQDNLNNDKKVVNFFVAKDGSTKEDQLVDALAAAPVAANFGVTLNSDGKPVDKDGKVLTGSDNDKNKLVSPAPIVLATDSLSSDQSVSISKVLDKDNGENLVQVGKGIATSVINKLKDLLS
使用单链 GT PDB 作为 Template 时,TMScore 是 0.4954,增加链间的 Docking 信息,TMScore 提升至 0.9917,提升很大。
逻辑位于 openfold/model/model.py,增加 globals.use_template_unmask 参数,修改逻辑:
if not self.globals.use_template_unmask:
multichain_mask_2d = (asym_id[..., None] == asym_id[..., None, :]) # [N_res, N_res]
else:
# 保留链内信息
# mask_inter = (asym_id[..., None] == asym_id[..., None, :])
# 保留链间信息
# mask_intra = (asym_id[..., None] != asym_id[..., None, :]) # [N_res, N_res]
# 保留全部信息
tmp_tensor = torch.ones(asym_id.shape, dtype=asym_id.dtype).to(asym_id.device)
mask_none = (tmp_tensor[..., None] == tmp_tensor[..., None, :])
mask_list = []
for i in range(n_tmpl):
# if i == 0:
# mask_list.append(mask_inter)
# else:
# mask_list.append(mask_intra)
mask_list.append(mask_none)
multichain_mask_2d = torch.stack(mask_list, dim=0)
logger.info(f"[CL] use_template_unmask: {self.globals.use_template_unmask}, "
f"multichain_mask_2d: {multichain_mask_2d.shape}")
# -------------- 验证 multichain_mask_2d -------------- #
# tmp = multichain_mask_2d.cpu().numpy()
# import pickle
# with open("multichain_mask_2d.pkl", "wb") as f:
# pickle.dump(tmp, f)
# logger.info(f"[CL] saved multichain_mask_2d!")
# -------------- 验证 multichain_mask_2d -------------- #
# 调用逻辑
template_embeds = self.template_embedder(
template_feats,
z,
pair_mask.to(dtype=z.dtype),
no_batch_dims,
chunk_size=self.globals.chunk_size,
multichain_mask_2d=multichain_mask_2d,
use_fa=self.globals.use_fa,
)
只保留链内信息:
multichain_mask_2d = (asym_id[..., None] == asym_id[..., None, :]) # [N_res, N_res]
只保留链间信息:
# 保留链间信息
multichain_mask_2d = (asym_id[..., None] != asym_id[..., None, :]) # [N_res, N_res]
保留全部(链内和链间)信息:
tmp_tensor = torch.ones(asym_id.shape, dtype=asym_id.dtype).to(asym_id.device)
multichain_mask_2d = (tmp_tensor[..., None] == tmp_tensor[..., None, :])
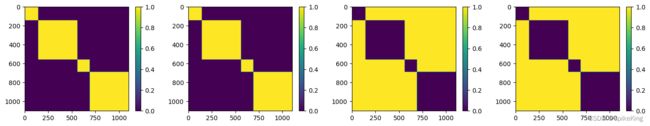
其中 multichain_mask_2d 特征,4 个特征,2 个链内 Mask,2 个链间 Mask,即:
其中,template_feats 特征维度,如下:
template_feats, template_all_atom_positions: torch.Size([4, 1102, 37, 3])
template_feats, template_all_atom_mask: torch.Size([4, 1102, 37])
template_feats, template_aatype: torch.Size([4, 1102])
template_feats, template_mask: torch.Size([4])
template_feats, template_pseudo_beta: torch.Size([4, 1102, 3])
template_feats, template_pseudo_beta_mask: torch.Size([4, 1102])
template_feats, template_torsion_angles_sin_cos: torch.Size([4, 1102, 7, 2])
template_feats, template_alt_torsion_angles_sin_cos: torch.Size([4, 1102, 7, 2])
template_feats, template_torsion_angles_mask: torch.Size([4, 1102, 7])
具体逻辑位于 openfold/model/embedders.py#TemplateEmbedderMultimer,即:
- 其中,循环处理多个 Template
for i in range(n_templ):
pair_act = self.template_pair_embedder(
template_dgram,
aatype_one_hot,
z,
pseudo_beta_mask,
backbone_mask,
multichain_mask_2d,
unit_vector,
)
调用 openfold/model/embedders.py#TemplatePairEmbedderMultimer,即:
# template_dgram 部分
pseudo_beta_mask_2d = pseudo_beta_mask_2d * multichain_mask_2d
template_dgram = template_dgram * pseudo_beta_mask_2d[..., None]
# ...
# rigid Rt 部分
backbone_mask_2d = backbone_mask_2d * multichain_mask_2d
x, y, z = [coord * backbone_mask_2d for coord in unit_vector]
# ...
使用 pickle 存储 Template 特征,即:
# 写入
if key == "template_all_atom_positions":
tmp = template_feats[key].cpu().numpy()
import pickle
with open("template_all_atom_positions.pkl", "wb") as f:
pickle.dump(tmp, f)
logger.info(f"[CL] saved template_all_atom_positions!")
# 读取
def load_tensor_pkl(input_path):
import pickle
with open(input_path, "rb") as f:
obj = pickle.load(f)
# print(f"[Info] feat_dict: {obj.keys()}")
return obj
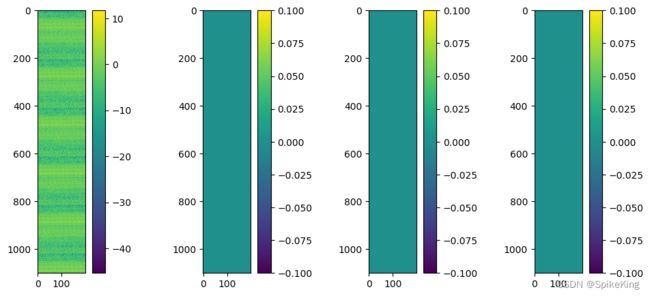
当只有 1组 Template 时,其他的模版 (例如最大模版数量是4) 填充至0,特征效果如下:
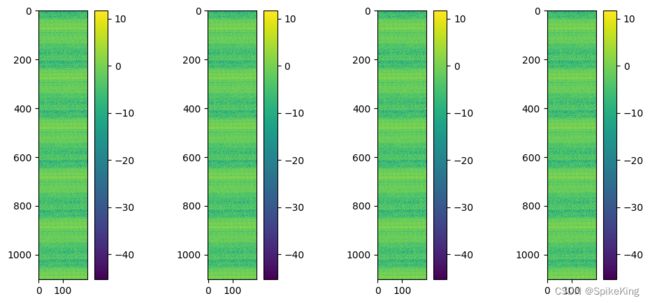
相同模版复制,特征效果如下,单模版效果0.9837,全模版效果0.9917: