mahout贝叶斯算法开发思路(拓展篇)2
如果想直接下面算法调用包,可以直接在mahout贝叶斯算法拓展下载,该算法调用的方式如下:
$HADOOP_HOME/bin hadoop jar mahout.jar mahout.fansy.bayes.BayerRunner -i hdfs_input_path -o hdfs_output_path -scl : -scv ,
调用参数如下:
usage: <command> [Generic Options] [Job-Specific Options]
Generic Options:
-archives <paths> comma separated archives to be unarchived
on the compute machines.
-conf <configuration file> specify an application configuration file
-D <property=value> use value for given property
-files <paths> comma separated files to be copied to the
map reduce cluster
-fs <local|namenode:port> specify a namenode
-jt <local|jobtracker:port> specify a job tracker
-libjars <paths> comma separated jar files to include in
the classpath.
-tokenCacheFile <tokensFile> name of the file with the tokens
Job-Specific Options:
--input (-i) input Path to job input
directory.
--output (-o) output The directory pathname
for output.
--splitCharacterVector (-scv) splitCharacterVector Vector split
character,default is
','
--splitCharacterLabel (-scl) splitCharacterLabel Vector and Label split
character,default is
':'
--help (-h) Print out help
--tempDir tempDir Intermediate output
directory
--startPhase startPhase First phase to run
--endPhase endPhase Last phase to run
接上篇分析下面的步骤:
4. 获取贝叶斯模型的属性值2:
这一步骤相当于 TrainNaiveBayesJob的第二个prepareJob,其中mapper和reducer都是参考这个job的,基本没有修改代码;代码如下:
package mahout.fansy.bayes;
import org.apache.hadoop.conf.Configuration;
import org.apache.hadoop.fs.Path;
import org.apache.hadoop.io.Text;
import org.apache.hadoop.mapreduce.Job;
import org.apache.hadoop.mapreduce.lib.input.SequenceFileInputFormat;
import org.apache.hadoop.mapreduce.lib.output.SequenceFileOutputFormat;
import org.apache.hadoop.util.ToolRunner;
import org.apache.mahout.classifier.naivebayes.training.WeightsMapper;
import org.apache.mahout.common.AbstractJob;
import org.apache.mahout.common.HadoopUtil;
import org.apache.mahout.common.mapreduce.VectorSumReducer;
import org.apache.mahout.math.VectorWritable;
/**
* 贝叶斯算法第二个job任务相当于 TrainNaiveBayesJob的第二个prepareJob
* Mapper,Reducer还用原来的
* @author Administrator
*
*/
public class BayesJob2 extends AbstractJob {
/**
* @param args
* @throws Exception
*/
public static void main(String[] args) throws Exception {
ToolRunner.run(new Configuration(), new BayesJob2(),args);
}
@Override
public int run(String[] args) throws Exception {
addInputOption();
addOutputOption();
addOption("labelNumber","ln", "The number of the labele ");
if (parseArguments(args) == null) {
return -1;
}
Path input = getInputPath();
Path output = getOutputPath();
String labelNumber=getOption("labelNumber");
Configuration conf=getConf();
conf.set(WeightsMapper.class.getName() + ".numLabels",labelNumber);
HadoopUtil.delete(conf, output);
Job job=new Job(conf);
job.setJobName("job2 get weightsFeture and weightsLabel by job1's output:"+input.toString());
job.setJarByClass(BayesJob2.class);
job.setInputFormatClass(SequenceFileInputFormat.class);
job.setOutputFormatClass(SequenceFileOutputFormat.class);
job.setMapperClass(WeightsMapper.class);
job.setMapOutputKeyClass(Text.class);
job.setMapOutputValueClass(VectorWritable.class);
job.setCombinerClass(VectorSumReducer.class);
job.setReducerClass(VectorSumReducer.class);
job.setOutputKeyClass(Text.class);
job.setOutputValueClass(VectorWritable.class);
SequenceFileInputFormat.setInputPaths(job, input);
SequenceFileOutputFormat.setOutputPath(job, output);
if(job.waitForCompletion(true)){
return 0;
}
return -1;
}
}
其单独调用方式如下:
usage: <command> [Generic Options] [Job-Specific Options]
Generic Options:
-archives <paths> comma separated archives to be unarchived
on the compute machines.
-conf <configuration file> specify an application configuration file
-D <property=value> use value for given property
-files <paths> comma separated files to be copied to the
map reduce cluster
-fs <local|namenode:port> specify a namenode
-jt <local|jobtracker:port> specify a job tracker
-libjars <paths> comma separated jar files to include in
the classpath.
-tokenCacheFile <tokensFile> name of the file with the tokens
Job-Specific Options:
--input (-i) input Path to job input directory.
--output (-o) output The directory pathname for output.
--labelNumber (-ln) labelNumber The number of the labele
--help (-h) Print out help
--tempDir tempDir Intermediate output directory
--startPhase startPhase First phase to run
--endPhase endPhase Last phase to run
其实也就是设置一个标识的个数而已,其他参考AbstractJob的默认参数;
5.贝叶斯模型写入文件:
这一步把3、4步骤的输出进行转换然后作为贝叶斯模型的一部分,然后把贝叶斯模型写入文件,其中的转换以及写入文件都参考BayesUtils中的相关方法,具体代码如下:
package mahout.fansy.bayes;
import java.io.IOException;
import mahout.fansy.bayes.util.OperateArgs;
import org.apache.commons.cli.ParseException;
import org.apache.hadoop.conf.Configuration;
import org.apache.hadoop.fs.Path;
import org.apache.hadoop.io.IntWritable;
import org.apache.hadoop.io.Text;
import org.apache.mahout.classifier.naivebayes.NaiveBayesModel;
import org.apache.mahout.classifier.naivebayes.training.ThetaMapper;
import org.apache.mahout.classifier.naivebayes.training.TrainNaiveBayesJob;
import org.apache.mahout.common.Pair;
import org.apache.mahout.common.iterator.sequencefile.PathFilters;
import org.apache.mahout.common.iterator.sequencefile.PathType;
import org.apache.mahout.common.iterator.sequencefile.SequenceFileDirIterable;
import org.apache.mahout.math.Matrix;
import org.apache.mahout.math.SparseMatrix;
import org.apache.mahout.math.Vector;
import org.apache.mahout.math.VectorWritable;
import com.google.common.base.Preconditions;
public class WriteBayesModel extends OperateArgs{
/**
* @param args,输入和输出都是没有用的,输入是job1和job 2 的输出,输出是model的路径
* model存储的路径是 输出路径下面的naiveBayesModel.bin文件
* @throws ParseException
* @throws IOException
*/
public static void main(String[] args) throws IOException, ParseException {
String[] arg={"-jt","ubuntu:9001",
"-i","",
"-o","",
"-mp","hdfs://ubuntu:9000/user/mahout/output_bayes/bayesModel",
"-bj1","hdfs://ubuntu:9000/user/mahout/output_bayes/job1",
"-bj2","hdfs://ubuntu:9000/user/mahout/output_bayes/job2"};
new WriteBayesModel().run(arg);
}
/**
* 把model写入文件中
* @param args
* @throws IOException
* @throws ParseException
*/
public int run(String[] args) throws IOException, ParseException{
// modelPath
setOption("mp","modelPath",true,"the path for bayesian model to store",true);
// bayes job 1 path
setOption("bj1","bayesJob1",true,"the path for bayes job 1",true);
// bayes job 2 path
setOption("bj2","bayesJob2",true,"the path for bayes job 2",true);
if(!parseArgs(args)){
return -1;
}
String job1Path=getNameValue("bj1");
String job2Path=getNameValue("bj2");
Configuration conf=getConf();
String modelPath=getNameValue("mp");
NaiveBayesModel naiveBayesModel=readFromPaths(job1Path,job2Path,conf);
naiveBayesModel.validate();
naiveBayesModel.serialize(new Path(modelPath), getConf());
System.out.println("Write bayesian model to '"+modelPath+"/naiveBayesModel.bin'");
return 0;
}
/**
* 摘自BayesUtils的readModelFromDir方法,只修改了相关路径
* @param job1Path
* @param job2Path
* @param conf
* @return
*/
public NaiveBayesModel readFromPaths(String job1Path,String job2Path,Configuration conf){
float alphaI = conf.getFloat(ThetaMapper.ALPHA_I, 1.0f);
// read feature sums and label sums
Vector scoresPerLabel = null;
Vector scoresPerFeature = null;
for (Pair<Text,VectorWritable> record : new SequenceFileDirIterable<Text, VectorWritable>(
new Path(job2Path), PathType.LIST, PathFilters.partFilter(), conf)) {
String key = record.getFirst().toString();
VectorWritable value = record.getSecond();
if (key.equals(TrainNaiveBayesJob.WEIGHTS_PER_FEATURE)) {
scoresPerFeature = value.get();
} else if (key.equals(TrainNaiveBayesJob.WEIGHTS_PER_LABEL)) {
scoresPerLabel = value.get();
}
}
Preconditions.checkNotNull(scoresPerFeature);
Preconditions.checkNotNull(scoresPerLabel);
Matrix scoresPerLabelAndFeature = new SparseMatrix(scoresPerLabel.size(), scoresPerFeature.size());
for (Pair<IntWritable,VectorWritable> entry : new SequenceFileDirIterable<IntWritable,VectorWritable>(
new Path(job1Path), PathType.LIST, PathFilters.partFilter(), conf)) {
scoresPerLabelAndFeature.assignRow(entry.getFirst().get(), entry.getSecond().get());
}
Vector perlabelThetaNormalizer = scoresPerLabel.like();
return new NaiveBayesModel(scoresPerLabelAndFeature, scoresPerFeature, scoresPerLabel, perlabelThetaNormalizer,
alphaI);
}
}
6. 应用贝叶斯模型分类原始数据:
这个部分的代码也基本是参考mahout中贝叶斯算法的源码,只是修改了其中的解析部分的代码而已,具体如下:
package mahout.fansy.bayes;
import java.io.IOException;
import org.apache.hadoop.conf.Configuration;
import org.apache.hadoop.fs.Path;
import org.apache.hadoop.io.Text;
import org.apache.hadoop.mapreduce.Job;
import org.apache.hadoop.mapreduce.Mapper;
import org.apache.hadoop.mapreduce.lib.input.SequenceFileInputFormat;
import org.apache.hadoop.mapreduce.lib.output.SequenceFileOutputFormat;
import org.apache.hadoop.util.ToolRunner;
import org.apache.mahout.classifier.naivebayes.AbstractNaiveBayesClassifier;
import org.apache.mahout.classifier.naivebayes.NaiveBayesModel;
import org.apache.mahout.classifier.naivebayes.StandardNaiveBayesClassifier;
import org.apache.mahout.classifier.naivebayes.training.WeightsMapper;
import org.apache.mahout.common.AbstractJob;
import org.apache.mahout.common.HadoopUtil;
import org.apache.mahout.math.Vector;
import org.apache.mahout.math.VectorWritable;
/**
* 用于分类的Job
* @author Administrator
*
*/
public class BayesClassifyJob extends AbstractJob {
/**
* @param args
* @throws Exception
*/
public static void main(String[] args) throws Exception {
ToolRunner.run(new Configuration(), new BayesClassifyJob(),args);
}
@Override
public int run(String[] args) throws Exception {
addInputOption();
addOutputOption();
addOption("model","m", "The file where bayesian model store ");
addOption("labelNumber","ln", "The labels number ");
if (parseArguments(args) == null) {
return -1;
}
Path input = getInputPath();
Path output = getOutputPath();
String labelNumber=getOption("labelNumber");
String modelPath=getOption("model");
Configuration conf=getConf();
conf.set(WeightsMapper.class.getName() + ".numLabels",labelNumber);
HadoopUtil.cacheFiles(new Path(modelPath), conf);
HadoopUtil.delete(conf, output);
Job job=new Job(conf);
job.setJobName("Use bayesian model to classify the input:"+input.getName());
job.setJarByClass(BayesClassifyJob.class);
job.setInputFormatClass(SequenceFileInputFormat.class);
job.setOutputFormatClass(SequenceFileOutputFormat.class);
job.setMapperClass(BayesClasifyMapper.class);
job.setMapOutputKeyClass(Text.class);
job.setMapOutputValueClass(VectorWritable.class);
job.setNumReduceTasks(0);
job.setOutputKeyClass(Text.class);
job.setOutputValueClass(VectorWritable.class);
SequenceFileInputFormat.setInputPaths(job, input);
SequenceFileOutputFormat.setOutputPath(job, output);
if(job.waitForCompletion(true)){
return 0;
}
return -1;
}
/**
* 自定义Mapper,只修改了解析部分代码
* @author Administrator
*
*/
public static class BayesClasifyMapper extends Mapper<Text, VectorWritable, Text, VectorWritable>{
private AbstractNaiveBayesClassifier classifier;
@Override
public void setup(Context context) throws IOException, InterruptedException {
System.out.println("Setup");
Configuration conf = context.getConfiguration();
Path modelPath = HadoopUtil.cachedFile(conf);
NaiveBayesModel model = NaiveBayesModel.materialize(modelPath, conf);
classifier = new StandardNaiveBayesClassifier(model);
}
@Override
public void map(Text key, VectorWritable value, Context context) throws IOException, InterruptedException {
Vector result = classifier.classifyFull(value.get());
//the key is the expected value
context.write(new Text(key.toString()), new VectorWritable(result));
}
}
}
如果要单独运行这一步,可以参考:
usage: <command> [Generic Options] [Job-Specific Options]
Generic Options:
-archives <paths> comma separated archives to be unarchived
on the compute machines.
-conf <configuration file> specify an application configuration file
-D <property=value> use value for given property
-files <paths> comma separated files to be copied to the
map reduce cluster
-fs <local|namenode:port> specify a namenode
-jt <local|jobtracker:port> specify a job tracker
-libjars <paths> comma separated jar files to include in
the classpath.
-tokenCacheFile <tokensFile> name of the file with the tokens
Job-Specific Options:
--input (-i) input Path to job input directory.
--output (-o) output The directory pathname for output.
--model (-m) model The file where bayesian model store
--labelNumber (-ln) labelNumber The labels number
--help (-h) Print out help
--tempDir tempDir Intermediate output directory
--startPhase startPhase First phase to run
--endPhase endPhase Last phase to run
只需提供model的路径和标识的个数这两个参数即可;
7. 对第6步分类的结果进行评价,这部分的代码如下:
package mahout.fansy.bayes;
import java.io.IOException;
import java.util.Map;
import mahout.fansy.bayes.util.OperateArgs;
import org.apache.commons.cli.ParseException;
import org.apache.hadoop.conf.Configuration;
import org.apache.hadoop.fs.Path;
import org.apache.hadoop.io.Text;
import org.apache.mahout.classifier.ClassifierResult;
import org.apache.mahout.classifier.ResultAnalyzer;
import org.apache.mahout.classifier.naivebayes.BayesUtils;
import org.apache.mahout.common.Pair;
import org.apache.mahout.common.iterator.sequencefile.PathFilters;
import org.apache.mahout.common.iterator.sequencefile.PathType;
import org.apache.mahout.common.iterator.sequencefile.SequenceFileDirIterable;
import org.apache.mahout.math.Vector;
import org.apache.mahout.math.VectorWritable;
import org.slf4j.Logger;
import org.slf4j.LoggerFactory;
public class AnalyzeBayesModel extends OperateArgs{
/**
* 输入是BayesClassifyJob的输出
* -o 参数没作用
*/
private static final Logger log = LoggerFactory.getLogger(AnalyzeBayesModel.class);
public static void main(String[] args) throws IOException, ParseException {
String[] arg={"-jt","ubuntu:9001",
"-i","hdfs://ubuntu:9000/user/mahout/output_bayes/classifyJob",
"-o","",
"-li","hdfs://ubuntu:9000/user/mahout/output_bayes/index.bin"
};
new AnalyzeBayesModel().run(arg);
}
/**
* 分析BayesClassifyJob输出文件和labelIndex做对比,分析正确率
* @param args
* @throws IOException
* @throws ParseException
*/
public int run(String[] args) throws IOException, ParseException{
// labelIndex
setOption("li","labelIndex",true,"the path where labelIndex store",true);
if(!parseArgs(args)){
return -1;
}
Configuration conf=getConf();
String labelIndex=getNameValue("labelIndex");
String input=getInput();
Path inputPath=new Path(input);
//load the labels
Map<Integer, String> labelMap = BayesUtils.readLabelIndex(getConf(), new Path(labelIndex));
//loop over the results and create the confusion matrix
SequenceFileDirIterable<Text, VectorWritable> dirIterable =
new SequenceFileDirIterable<Text, VectorWritable>(inputPath,
PathType.LIST,
PathFilters.partFilter(),
conf);
ResultAnalyzer analyzer = new ResultAnalyzer(labelMap.values(), "DEFAULT");
analyzeResults(labelMap, dirIterable, analyzer);
log.info("{} Results: {}", "Standard NB", analyzer);
return 0;
}
/**
* 摘自TestNaiveBayesDriver中的analyzeResults方法
*/
private void analyzeResults(Map<Integer, String> labelMap,
SequenceFileDirIterable<Text, VectorWritable> dirIterable,
ResultAnalyzer analyzer) {
for (Pair<Text, VectorWritable> pair : dirIterable) {
int bestIdx = Integer.MIN_VALUE;
double bestScore = Long.MIN_VALUE;
for (Vector.Element element : pair.getSecond().get()) {
if (element.get() > bestScore) {
bestScore = element.get();
bestIdx = element.index();
}
}
if (bestIdx != Integer.MIN_VALUE) {
ClassifierResult classifierResult = new ClassifierResult(labelMap.get(bestIdx), bestScore);
analyzer.addInstance(pair.getFirst().toString(), classifierResult);
}
}
}
}
运行拓展篇1中的数据得到的模型的分类结果如下:
13/09/14 14:52:13 INFO bayes.AnalyzeBayesModel: Standard NB Results: ======================================================= Summary ------------------------------------------------------- Correctly Classified Instances : 7 70% Incorrectly Classified Instances : 3 30% Total Classified Instances : 10 ======================================================= Confusion Matrix ------------------------------------------------------- a b c d <--Classified as 3 0 0 0 | 3 a = 1 0 1 0 1 | 2 b = 2 1 1 2 0 | 4 c = 3 0 0 0 1 | 1 d = 4
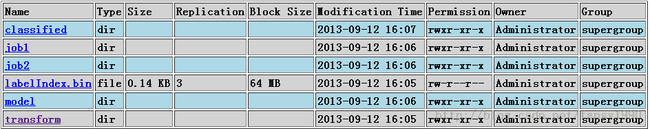
运行后可以在hdfs上面看到如下的文件夹:
分享,成长,快乐
转载请注明blog地址:http://blog.csdn.net/fansy1990