- leetcode-124 Binary Tree Maximum Path Sum
乐观的大鹏
LeetCode
Givenanon-emptybinarytree,findthemaximumpathsum.Forthisproblem,apathisdefinedasanysequenceofnodesfromsomestartingnodetoanynodeinthetreealongtheparent-childconnections.Thepathmustcontainatleastonenodea
- LeetCode 673. Number of Longest Increasing Subsequence (Java版; Meidum)
littlehaes
字符串动态规划算法leetcode数据结构
welcometomyblogLeetCode673.NumberofLongestIncreasingSubsequence(Java版;Meidum)题目描述Givenanunsortedarrayofintegers,findthenumberoflongestincreasingsubsequence.Example1:Input:[1,3,5,4,7]Output:2Explanatio
- Biopython提取和分离复合体PDB文件中所有链的结构信息
qq_27390023
生物信息学python
从蛋白质复合体的PDB文件中提取每个链的结构信息,并保存成单独的pdb文件。示例代码fromBioimportPDBdefextract_chain_sequences(pdb_file,output_dir):"""从PDB文件中提取所有链的序列,并保存为独立的PDB文件。:parampdb_file:蛋白质复合体PDB文件路径:paramoutput_dir:输出目录,用于保存各链的PDB文件
- 详解TCP的三次握手
汪先声
tcp/ip网络协议网络
TCP(三次握手)是指在建立一个可靠的传输控制协议(TCP)连接时,客户端和服务器之间的三步交互过程。这个过程的主要目的是确保连接是可靠的、双方的发送与接收能力是正常的,并且可以开始数据传输。下面是对每个步骤的详细解释:1.第一次握手:客户端发送SYN过程:客户端(A)向服务器(B)发送一个同步报文段(SYN,SynchronizeSequenceNumber),表示它想要与服务器建立连接。目的:
- LeetCode 2207. 字符串中最多数目的子字符串
Sasakihaise_
LeetCodeleetcode后缀和
题目链接:力扣https://leetcode-cn.com/problems/maximize-number-of-subsequences-in-a-string/【分析】由于pattern中只有两个字符,假设分别是a、b,只需要统计出text中每个a后面有多少b即可,这儿这个通过后缀和的思想,先算出总的b的个数,如果当前字符是a,那么后面b的个数就是总的b的个数,如果是b,就把总的b的个数-
- 自学Python:计算斐波纳契数列
小强聊成长
斐波那契数列(Fibonaccisequence),又称黄金分割数列,因数学家莱昂纳多·斐波那契(LeonardodaFibonacci)以兔子繁殖为例子而引入,故又称为“兔子数列”,指的是这样一个数列:0、1、1、2、3、5、8、13、21、34、……在数学上,斐波那契数列以如下被以递推的方法定义:F(0)=0,F(1)=1,F(n)=F(n-1)+F(n-2)(n≥2,n∈N*)在现代物理、准
- 一维数组 list 呢 ,怎么转换成 (批次 句子长度 特征值 )三维向量 python pytorch lstm 编程 人工智能
zhangfeng1133
pythonpytorch人工智能数据挖掘
一、介绍对于一维数组,如果你想将其转换成适合深度学习模型(如LSTM)输入的格式,你需要考虑将其扩展为三维张量。这通常涉及到批次大小(batchsize)、序列长度(sequencelength)和特征数量(numberoffeatures)的维度。以下是如何将一维数组转换为这种格式的步骤:###1.确定维度-**批次大小(BatchSize)**:这是你一次处理的样本数量。-**序列长度(Seq
- 顺序表(SequenceList)数据结构的基本操作实现详解
SuchABigBug
C语言数据结构链表c语言
目录一、前言二、整体设计框架三、函数实现1.SeqListInit2.SeqListDestory3.SeqListCheckCapacity4.SeqListPushBack(尾插)5.SeqListPopBack(尾删)6.SeqListPushFront(头插)7.SeqListPopFront(头删)8.SeqlistFind9.SeqListInsert(任意位置插入)9.SeqList
- 动态规划算法之最长公子序列详细解读(附带Java代码解读)
南城花随雪。
算法分析算法动态规划java
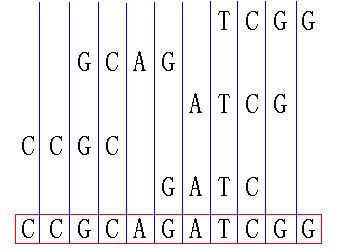
最长公共子序列(LongestCommonSubsequence,LCS)问题是动态规划中另一个经典问题,广泛用于比较两个序列的相似度。它的目标是找到两个序列之间最长的公共子序列(不是连续的),使得这个子序列同时出现在两个序列中。1.问题定义给定两个序列X和Y,要找到它们的最长公共子序列,即一个序列Z,它同时是X和Y的子序列,且Z的长度最大。例如:对于序列X="ABCBDAB"和Y="BDCAB"
- LeetCode题解:Word Ladder
CheeRok
LeetCodeLeetCode全题解leetcode
Giventwowords(beginWordandendWord),andadictionary’swordlist,findthelengthofshortesttransformationsequencefrombeginWordtoendWord,suchthat:OnlyonelettercanbechangedatatimeEachintermediatewordmustexistin
- C语言 | Leetcode C语言题解之第392题判断子序列
DdddJMs__135
分享C语言Leetcode题解
题目:题解:boolisSubsequence(char*s,char*t){intn=strlen(s),m=strlen(t);intf[m+1][26];memset(f,0,sizeof(f));for(inti=0;i=0;i--){for(intj=0;j<26;j++){if(t[i]==j+'a')f[i][j]=i;elsef[i][j]=f[i+1][j];}}intadd=0
- 代码随想录算法训练营Day22 | 491.递增子序列,46.全排列,47.全排列 II ,332. 重新安排行程,51. N皇后,37. 解数独,总结
Yummy Penguin
算法
第七章回溯算法part04491.递增子序列本题和大家刚做过的90.子集II非常像,但又很不一样,很容易掉坑里。代码随想录视频讲解:回溯算法精讲,树层去重与树枝去重|LeetCode:491.递增子序列_哔哩哔哩_bilibili#491classSolution:deffindSubsequences(self,nums):result=[]path=[]self.backtracking(nu
- C++ | Leetcode C++题解之第392题判断子序列
Ddddddd_158
经验分享C++Leetcode题解
题目:题解:classSolution{public:boolisSubsequence(strings,stringt){intn=s.size(),m=t.size();vector>f(m+1,vector(26,0));for(inti=0;i=0;i--){for(intj=0;j<26;j++){if(t[i]==j+'a')f[i][j]=i;elsef[i][j]=f[i+1][j
- python安全渗透笔记
红云谈安全
python编程学习python安全
Python学习python常见错误UnicodeDecodeError:‘gbk’codeccan’tdecodebyte0x8cinposition22:illegalmultibytesequence//打开的文件未编码encoding='UTF-8'expectedanindentedblock未缩进,ifforcontinue又进入一次新的循环%将其他变量置入字符串特定位置以生成新字符串
- 胶水105
假装是老付
捕捉这些脱节的最好方法是按顺序给我自己讲一个故事,并考虑到我从以前的那些对话中收集到【工作任务之间】的那些依赖关系。Thebestwaytocatchthesedisconnectsistotellmyselfastoryinasequence,consideringdependenciesI’vegleanedfrompreviousconversations.例二EXAMPLE2来自销售团队的
- Kafka高性能揭秘 —— sequence IO、PageCache、SendFile的应用详解
大数据学习与分享
Kafkakafka大数据java
大家都知道Kafka是将数据存储于磁盘的,而磁盘读写性能往往很差,但Kafka官方测试其数据读写速率能达到600M/s,那么为什么Kafka性能会这么高呢?首先producer往broker发送消息时,采用batch的方式即批量而非一条一条的发送,这种方式可以有效降低网络IO的请求次数,提升性能。此外这些批次消息会"暂存"在缓冲池中,避免频繁的GC问题。批量发送的消息可以进行压缩并且传输的时候可以
- vLLM (3) - Sequence & SequenceGroup
戴昊光
人工智能languagemodelnlppythontransformer
系列文章目录vLLM(1)-Qwen2推理&部署vLLM(2)-架构总览vLLM(3)-Sequence&SequenceGroup文章目录系列文章目录前言一、SequenceStage&SequenceStatus1.SeqenceStage2.SeqenceStatus二、SequenceData&Sequence1.SequenceData2.Sequence三、SequenceGroup&
- 创建sequence sql模板
wrx繁星点点
#新增模板sql数据库
createsequenceSEQ_CLE_FUND_ADJU_APPLY(序列名)minvalue(最小值)1nomaxvalue(如果间隔值>0说明是递增序列可以不需要最大值就使用nomaxvalue代替maxvalue9999999999,如果间隔值<0说明是递减序列可以不需要最小值就可以使用nominvalue)startwith(起始值:默认为1)1incrementby(间隔数:每次加
- 浙大数据结构:01-复杂度2 Maximum Subsequence Sum
_Power_Y
数据结构浙大数据结构c++
数据结构MOOCPTA习题01-复杂度2MaximumSubsequenceSum#includeusingnamespacestd;constintM=100005;inta[M];intmain(){intk;cin>>k;intf=1;for(inti=0;i>a[i];if(a[i]>=0)//如果出现大于0则进行在线处理f=0;}if(f){//全都小于0coutma){//更新答案ma
- hive序列生成_Hive实现自增列的两种方法
weixin_39559804
hive序列生成
多维数据仓库中的维度表和事实表一般都需要有一个代理键,作为这些表的主键,代理键一般由单列的自增数字序列构成。Hive没有关系数据库中的自增列,但它也有一些对自增序列的支持,通常有两种方法生成代理键:使用row_number()窗口函数或者使用一个名为UDFRowSequence的用户自定义函数(UDF)。用row_number()函数生成代理键INSERTOVERWRITETABLEmy_hive
- 2020-08-13 To Succeed in The Long-Term, Think in 5, 10, or 20 Year Time Frames
春生阁
Humanscanbeprettygoodatshort-termthinking—butwe’renotverygoodatthinkingaboutthelong-termconsequencesofourpresentactions.Thepressuresofmodernlifemakeitincrediblydifficultforustoprojecttheimplicationsof
- AtCoder Beginner Contest 369(ABCDEFG题)视频讲解
阿史大杯茶
Atcoderc++算法数据结构
A-369ProblemStatementYouaregiventwointegersAAAandBBB.Howmanyintegersxxxsatisfythefollowingcondition?Condition:ItispossibletoarrangethethreeintegersAAA,BBB,andxxxinsomeordertoformanarithmeticsequence.A
- Java | Leetcode Java题解之第392题判断子序列
m0_57195758
分享JavaLeetcode题解
题目:题解:classSolution{publicbooleanisSubsequence(Strings,Stringt){intn=s.length(),m=t.length();int[][]f=newint[m+1][26];for(inti=0;i=0;i--){for(intj=0;j<26;j++){if(t.charAt(i)==j+'a')f[i][j]=i;elsef[i][
- C++ | Leetcode C++题解之第392题判断子序列
Ddddddd_158
经验分享C++Leetcode题解
题目:题解:classSolution{public:boolisSubsequence(strings,stringt){intn=s.size(),m=t.size();vector>f(m+1,vector(26,0));for(inti=0;i=0;i--){for(intj=0;j<26;j++){if(t[i]==j+'a')f[i][j]=i;elsef[i][j]=f[i+1][j
- 如何在oracle实现自增数列
bhots
oracle数据库
如果您想在Oracle数据库中使用触发器(Trigger)实现自增功能,可以考虑通过序列(Sequence)结合触发器实现自增功能。以下是一个简单的示例:创建序列:首先创建一个序列,用于生成自增的值。CREATESEQUENCEyour_sequence_nameSTARTWITH1INCREMENTBY1;创建触发器:创建一个触发器,在插入数据时触发,将序列的值插入到相应的列中。CREATEOR
- 斐波那契数列——C语言
木木ᶻ
c语言蓝桥杯算法
目录一、递归法二、for循环三、for循环+数组斐波那契数列(Fibonaccisequence),也称之为黄金分割数列,由意大利数学家列昂纳多・斐波那契(LeonardoFibonacci)提出。斐波那契数列指的是这样的一个数列:1、1、2、3、5、8、13、21、34、……,这个数列从第3项开始,每一项都等于前面两项之和。在数学上,斐波那契数列可以被递推的方法定义如下:F(1)=1F(2)=1
- Hive的存储格式
百流
hadoop学习日记hivehadoop数据仓库
文章目录Hive的存储格式1.存储格式简介2.行存储与列存储行式存储列式存储混合的PAX存储结构TextFileSequenceFileHive的存储格式1.存储格式简介Hive支持的存储数的格式主要有:TEXTFILE(默认格式)、SEQUENCEFILE、RCFILE、ORCFILE、PARQUET。textfile为默认格式,建表时没有指定文件格式,则使用TEXTFILE,导入数据时会直接把
- PCIe - DMA Sequence
Starry丶
标准总线接口协议数字ICfpga开发express网络
目录1.Initiation2.H2C(HosttoCard)2.1.MWr2.2.MRd2.3.CfgWr2.4.CfgRd3.C2H(CardtoHost)3.1.MWr3.2.MRd介绍PCIe拓扑结构下的DMA流程,实际上RC充当的就是DMAController的角色1.Initiation上电初始化,在设备枚举阶段,在内存空间中为每个PCIe设备分配BAR空间(包括MSI-X中断向量表)
- oracle创建的用户能看另一用户全部表,ORACLE授权用户查询另一个用户下的表与视图...
实在想不出来了
一、系统权限说明:1、用户权限CREATESESSIOIN连接到数据库CREATETABLE在用户的方案中创建表CREATESEQUENCE在用户的方案中创建序列CREATEVIEW在用户的方案中创视图CREATEPROCEDURE在用户的方案中创建存储过程,函数或包1.1、例子:授予系统权限DBA能够授予用户指定的系统权限GRANTcreatesession,createtable,create
- QT-快捷键-记事本文字放大缩小
打地基的小白
QT学习路程qt开发语言
一、QShortcut类QShortcut简介QShortcut类允许你为你的应用程序定义快捷键。当快捷键被按下时,QShortcut可以触发一个槽函数,或者它可以直接激活一个QWidget。创建QShortcut对象QShortcut可以通过多种构造函数创建,其中最常见的形式是:QShortcut(QKeySequencekey,QWidget*parent,constchar*member=0
- 312个免费高速HTTP代理IP(能隐藏自己真实IP地址)
yangshangchuan
高速免费superwordHTTP代理
124.88.67.20:843
190.36.223.93:8080
117.147.221.38:8123
122.228.92.103:3128
183.247.211.159:8123
124.88.67.35:81
112.18.51.167:8123
218.28.96.39:3128
49.94.160.198:3128
183.20
- pull解析和json编码
百合不是茶
androidpull解析json
n.json文件:
[{name:java,lan:c++,age:17},{name:android,lan:java,age:8}]
pull.xml文件
<?xml version="1.0" encoding="utf-8"?>
<stu>
<name>java
- [能源与矿产]石油与地球生态系统
comsci
能源
按照苏联的科学界的说法,石油并非是远古的生物残骸的演变产物,而是一种可以由某些特殊地质结构和物理条件生产出来的东西,也就是说,石油是可以自增长的....
那么我们做一个猜想: 石油好像是地球的体液,我们地球具有自动产生石油的某种机制,只要我们不过量开采石油,并保护好
- 类与对象浅谈
沐刃青蛟
java基础
类,字面理解,便是同一种事物的总称,比如人类,是对世界上所有人的一个总称。而对象,便是类的具体化,实例化,是一个具体事物,比如张飞这个人,就是人类的一个对象。但要注意的是:张飞这个人是对象,而不是张飞,张飞只是他这个人的名字,是他的属性而已。而一个类中包含了属性和方法这两兄弟,他们分别用来描述对象的行为和性质(感觉应该是
- 新站开始被收录后,我们应该做什么?
IT独行者
PHPseo
新站开始被收录后,我们应该做什么?
百度终于开始收录自己的网站了,作为站长,你是不是觉得那一刻很有成就感呢,同时,你是不是又很茫然,不知道下一步该做什么了?至少我当初就是这样,在这里和大家一份分享一下新站收录后,我们要做哪些工作。
至于如何让百度快速收录自己的网站,可以参考我之前的帖子《新站让百
- oracle 连接碰到的问题
文强chu
oracle
Unable to find a java Virtual Machine--安装64位版Oracle11gR2后无法启动SQLDeveloper的解决方案
作者:草根IT网 来源:未知 人气:813标签:
导读:安装64位版Oracle11gR2后发现启动SQLDeveloper时弹出配置java.exe的路径,找到Oracle自带java.exe后产生的路径“C:\app\用户名\prod
- Swing中按ctrl键同时移动鼠标拖动组件(类中多借口共享同一数据)
小桔子
java继承swing接口监听
都知道java中类只能单继承,但可以实现多个接口,但我发现实现多个接口之后,多个接口却不能共享同一个数据,应用开发中想实现:当用户按着ctrl键时,可以用鼠标点击拖动组件,比如说文本框。
编写一个监听实现KeyListener,NouseListener,MouseMotionListener三个接口,重写方法。定义一个全局变量boolea
- linux常用的命令
aichenglong
linux常用命令
1 startx切换到图形化界面
2 man命令:查看帮助信息
man 需要查看的命令,man命令提供了大量的帮助信息,一般可以分成4个部分
name:对命令的简单说明
synopsis:命令的使用格式说明
description:命令的详细说明信息
options:命令的各项说明
3 date:显示时间
语法:date [OPTION]... [+FORMAT]
- eclipse内存优化
AILIKES
javaeclipsejvmjdk
一 基本说明 在JVM中,总体上分2块内存区,默认空余堆内存小于 40%时,JVM就会增大堆直到-Xmx的最大限制;空余堆内存大于70%时,JVM会减少堆直到-Xms的最小限制。 1)堆内存(Heap memory):堆是运行时数据区域,所有类实例和数组的内存均从此处分配,是Java代码可及的内存,是留给开发人
- 关键字的使用探讨
百合不是茶
关键字
//关键字的使用探讨/*访问关键词private 只能在本类中访问public 只能在本工程中访问protected 只能在包中和子类中访问默认的 只能在包中访问*//*final 类 方法 变量 final 类 不能被继承 final 方法 不能被子类覆盖,但可以继承 final 变量 只能有一次赋值,赋值后不能改变 final 不能用来修饰构造方法*///this()
- JS中定义对象的几种方式
bijian1013
js
1. 基于已有对象扩充其对象和方法(只适合于临时的生成一个对象):
<html>
<head>
<title>基于已有对象扩充其对象和方法(只适合于临时的生成一个对象)</title>
</head>
<script>
var obj = new Object();
- 表驱动法实例
bijian1013
java表驱动法TDD
获得月的天数是典型的直接访问驱动表方式的实例,下面我们来展示一下:
MonthDaysTest.java
package com.study.test;
import org.junit.Assert;
import org.junit.Test;
import com.study.MonthDays;
public class MonthDaysTest {
@T
- LInux启停重启常用服务器的脚本
bit1129
linux
启动,停止和重启常用服务器的Bash脚本,对于每个服务器,需要根据实际的安装路径做相应的修改
#! /bin/bash
Servers=(Apache2, Nginx, Resin, Tomcat, Couchbase, SVN, ActiveMQ, Mongo);
Ops=(Start, Stop, Restart);
currentDir=$(pwd);
echo
- 【HBase六】REST操作HBase
bit1129
hbase
HBase提供了REST风格的服务方便查看HBase集群的信息,以及执行增删改查操作
1. 启动和停止HBase REST 服务 1.1 启动REST服务
前台启动(默认端口号8080)
[hadoop@hadoop bin]$ ./hbase rest start
后台启动
hbase-daemon.sh start rest
启动时指定
- 大话zabbix 3.0设计假设
ronin47
What’s new in Zabbix 2.0?
去年开始使用Zabbix的时候,是1.8.X的版本,今年Zabbix已经跨入了2.0的时代。看了2.0的release notes,和performance相关的有下面几个:
:: Performance improvements::Trigger related da
- http错误码大全
byalias
http协议javaweb
响应码由三位十进制数字组成,它们出现在由HTTP服务器发送的响应的第一行。
响应码分五种类型,由它们的第一位数字表示:
1)1xx:信息,请求收到,继续处理
2)2xx:成功,行为被成功地接受、理解和采纳
3)3xx:重定向,为了完成请求,必须进一步执行的动作
4)4xx:客户端错误,请求包含语法错误或者请求无法实现
5)5xx:服务器错误,服务器不能实现一种明显无效的请求
- J2EE设计模式-Intercepting Filter
bylijinnan
java设计模式数据结构
Intercepting Filter类似于职责链模式
有两种实现
其中一种是Filter之间没有联系,全部Filter都存放在FilterChain中,由FilterChain来有序或无序地把把所有Filter调用一遍。没有用到链表这种数据结构。示例如下:
package com.ljn.filter.custom;
import java.util.ArrayList;
- 修改jboss端口
chicony
jboss
修改jboss端口
%JBOSS_HOME%\server\{服务实例名}\conf\bindingservice.beans\META-INF\bindings-jboss-beans.xml
中找到
<!-- The ports-default bindings are obtained by taking the base bindin
- c++ 用类模版实现数组类
CrazyMizzz
C++
最近c++学到数组类,写了代码将他实现,基本具有vector类的功能
#include<iostream>
#include<string>
#include<cassert>
using namespace std;
template<class T>
class Array
{
public:
//构造函数
- hadoop dfs.datanode.du.reserved 预留空间配置方法
daizj
hadoop预留空间
对于datanode配置预留空间的方法 为:在hdfs-site.xml添加如下配置
<property>
<name>dfs.datanode.du.reserved</name>
<value>10737418240</value>
- mysql远程访问的设置
dcj3sjt126com
mysql防火墙
第一步: 激活网络设置 你需要编辑mysql配置文件my.cnf. 通常状况,my.cnf放置于在以下目录: /etc/mysql/my.cnf (Debian linux) /etc/my.cnf (Red Hat Linux/Fedora Linux) /var/db/mysql/my.cnf (FreeBSD) 然后用vi编辑my.cnf,修改内容从以下行: [mysqld] 你所需要: 1
- ios 使用特定的popToViewController返回到相应的Controller
dcj3sjt126com
controller
1、取navigationCtroller中的Controllers
NSArray * ctrlArray = self.navigationController.viewControllers;
2、取出后,执行,
[self.navigationController popToViewController:[ctrlArray objectAtIndex:0] animated:YES
- Linux正则表达式和通配符的区别
eksliang
正则表达式通配符和正则表达式的区别通配符
转载请出自出处:http://eksliang.iteye.com/blog/1976579
首先得明白二者是截然不同的
通配符只能用在shell命令中,用来处理字符串的的匹配。
判断一个命令是否为bash shell(linux 默认的shell)的内置命令
type -t commad
返回结果含义
file 表示为外部命令
alias 表示该
- Ubuntu Mysql Install and CONF
gengzg
Install
http://www.navicat.com.cn/download/navicat-for-mysql
Step1: 下载Navicat ,网址:http://www.navicat.com/en/download/download.html
Step2:进入下载目录,解压压缩包:tar -zxvf navicat11_mysql_en.tar.gz
- 批处理,删除文件bat
huqiji
windowsdos
@echo off
::演示:删除指定路径下指定天数之前(以文件名中包含的日期字符串为准)的文件。
::如果演示结果无误,把del前面的echo去掉,即可实现真正删除。
::本例假设文件名中包含的日期字符串(比如:bak-2009-12-25.log)
rem 指定待删除文件的存放路径
set SrcDir=C:/Test/BatHome
rem 指定天数
set DaysAgo=1
- 跨浏览器兼容的HTML5视频音频播放器
天梯梦
html5
HTML5的video和audio标签是用来在网页中加入视频和音频的标签,在支持html5的浏览器中不需要预先加载Adobe Flash浏览器插件就能轻松快速的播放视频和音频文件。而html5media.js可以在不支持html5的浏览器上使video和audio标签生效。 How to enable <video> and <audio> tags in
- Bundle自定义数据传递
hm4123660
androidSerializable自定义数据传递BundleParcelable
我们都知道Bundle可能过put****()方法添加各种基本类型的数据,Intent也可以通过putExtras(Bundle)将数据添加进去,然后通过startActivity()跳到下一下Activity的时候就把数据也传到下一个Activity了。如传递一个字符串到下一个Activity
把数据放到Intent
- C#:异步编程和线程的使用(.NET 4.5 )
powertoolsteam
.net线程C#异步编程
异步编程和线程处理是并发或并行编程非常重要的功能特征。为了实现异步编程,可使用线程也可以不用。将异步与线程同时讲,将有助于我们更好的理解它们的特征。
本文中涉及关键知识点
1. 异步编程
2. 线程的使用
3. 基于任务的异步模式
4. 并行编程
5. 总结
异步编程
什么是异步操作?异步操作是指某些操作能够独立运行,不依赖主流程或主其他处理流程。通常情况下,C#程序
- spark 查看 job history 日志
Stark_Summer
日志sparkhistoryjob
SPARK_HOME/conf 下:
spark-defaults.conf 增加如下内容
spark.eventLog.enabled true spark.eventLog.dir hdfs://master:8020/var/log/spark spark.eventLog.compress true
spark-env.sh 增加如下内容
export SP
- SSH框架搭建
wangxiukai2015eye
springHibernatestruts
MyEclipse搭建SSH框架 Struts Spring Hibernate
1、new一个web project。
2、右键项目,为项目添加Struts支持。
选择Struts2 Core Libraries -<MyEclipes-Library>
点击Finish。src目录下多了struts