简单易学的机器学习算法——谱聚类(Spectal Clustering)
一、复杂网络中的一些基本概念
1、复杂网络的表示
在复杂网络的表示中,复杂网络可以建模成一个图,其中, 表示网络中的节点的集合,
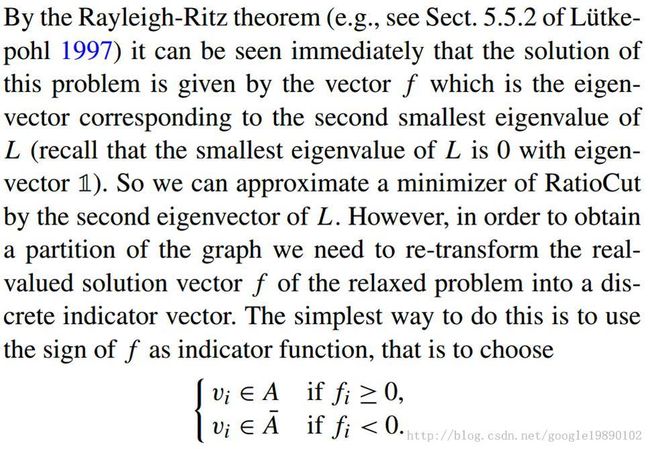
表示网络中的节点的集合,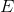 表示的是连接的集合。在复杂网络中,复杂网络可以是无向图、有向图、加权图或者超图。
表示的是连接的集合。在复杂网络中,复杂网络可以是无向图、有向图、加权图或者超图。
2、网络簇结构
网络簇结构
(network cluster structure)
也称为网络社团结构
(network community structure)
,是复杂网络中最普遍和最重要的拓扑属性之一。网络簇是整个网络中的稠密连接分支,具有同簇内部节点之间相互连接密集,不同簇的节点之间相互连接稀疏的特征。
3、复杂网络的分类
复杂网络主要分为:随机网络,小世界网络和无标度网络。
二、谱方法介绍
1、谱方法的思想
在复杂网络的网络簇结构存在着同簇节点之间连接密集,不同簇节点之间连接稀疏的特征,是否可以根据这样的特征对网络中的节点进行聚类,使得同类节点之间的连接密集,不同类别节点之间的连接稀疏?
在谱聚类中定义了“截”函数的概念,当一个网络被划分成为两个子网络时,“截”即指子网间的连接密度。谱聚类的目的就是要找到一种合理的分割,使得分割后形成若干子图,连接不同的子图的边的权重尽可能低,即“截”最小,同子图内的边的权重尽可能高。
2、“截”函数的具体表现形式
“截”表示的是子网间的密度,即边比较少。以二分为例,将图聚类成两个类: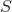 类和类。假设用
类和类。假设用 来表示图的划分,我们需要的结果为:
来表示图的划分,我们需要的结果为:
其中表示的是类别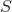 和之间的权重。
对于
和之间的权重。
对于
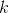 个不同的类别
,优化的目标为:
个不同的类别
,优化的目标为:
3、基本“截”函数的弊端
对于上述的“截”函数,最终会导致不好的分割,如二分类问题:
上述的“截”函数通常会将图分割成一个点和其余个点。
4、其他的“截”函数的表现形式
为了能够让每个类都有合理的大小,目标函数中应该使得足够大,则提出了 或者:
或者:
其中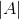 表示
表示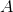 类中包含的顶点的数目
类中包含的顶点的数目
三、Laplacian矩阵
1、Laplacian矩阵的定义
拉普拉斯矩阵
(Laplacian Matrix)
,也称为基尔霍夫矩阵,是图的一种矩阵表示形式。
对于一个有 个顶点的图,其
Laplacian
矩阵定义为:
个顶点的图,其
Laplacian
矩阵定义为:
其中, 为图的度矩阵,
为图的度矩阵,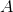 为图的邻接矩阵。
为图的邻接矩阵。
2、度矩阵的定义
度矩阵是一个对角矩阵,主角线上的值由对应的顶点的度组成。
对于一个有 个顶点的图,其邻接矩阵为:
个顶点的图,其邻接矩阵为:
其度矩阵为:
3、Laplacian矩阵的性质
- Laplacian矩阵
 是对称半正定矩阵;
是对称半正定矩阵; - Laplacian矩阵
 的最小特征值是
的最小特征值是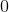 ,相应的特征向量是;
,相应的特征向量是; - Laplacian矩阵
 有
有 个非负实特征值:
个非负实特征值: ,且对于任何一个实向量
,且对于任何一个实向量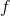 ,都有下面的式子成立:
,都有下面的式子成立:
性质3的证明:
4、不同的Laplacian矩阵
除了上述的拉普拉斯矩阵,还有规范化的
Laplacian
矩阵形式:
四、Laplacian矩阵与谱聚类中的优化函数的关系
1、由Laplacian矩阵到“截”函数
对于二个类别的聚类问题,优化的目标函数为:
定义向量,且
而已知:,则
而![\frac{1}{2}\cdot \left [ \sum_{i\in A,j\in \bar{A}}w_{ij}+\sum_{i\in \bar{A},j\in A}w_{ij} \right ]=\frac{1}{2}\sum_{i=1}^{k}W\left ( A_i,\bar{A}_i \right )=cut\left ( A,\bar{A} \right )](http://latex.codecogs.com/gif.latex?\frac{1}{2}\cdot&space;\left&space;[&space;\sum_{i\in&space;A,j\in&space;\bar{A}}w_{ij}+\sum_{i\in&space;\bar{A},j\in&space;A}w_{ij}&space;\right&space;]=\frac{1}{2}\sum_{i=1}^{k}W\left&space;(&space;A_i,\bar{A}_i&space;\right&space;)=cut\left&space;(&space;A,\bar{A}&space;\right&space;))
而}{\left&space;|&space;A&space;\right&space;|}+\frac{cut\left&space;(&space;A,\bar{A}&space;\right&space;)}{\left&space;|&space;\bar{A}&space;\right&space;|}&space;\right&space;)=\sum_{i=1}^{k}\frac{cut\left&space;(&space;A_i,\bar{A}_i&space;\right&space;)}{\left&space;|&space;A_i&space;\right&space;|}=RatioCut\left&space;(&space;A,\bar{A}&space;\right&space;))
其中,表示的是顶点的数目,对于确定的图来说是个常数。由上述的推导可知,由推导出了,由此可知:
Laplacian
矩阵与有优化的目标函数之间存在密切的联系。
2、新的目标函数
由上式可得:
由于是个常数,故要求的最小值,即求的最小值。则新的目标函数为:
其中
3、转化到Laplacian矩阵的求解
假设 是
Laplacian
矩阵
是
Laplacian
矩阵 的特征值,
的特征值,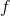 是特征值
是特征值 对应的特征向量,则有:
对应的特征向量,则有:
在上式的两端同时左乘
已知,则,上式可以转化为:
要求 ,即只需求得最小特征值
,即只需求得最小特征值 。由
Laplacian
矩阵的性质可知,
Laplacian
矩阵的最小特征值为
。由
Laplacian
矩阵的性质可知,
Laplacian
矩阵的最小特征值为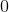 。由
Rayleigh-Ritz
理论,可以取第2小特征值。
。由
Rayleigh-Ritz
理论,可以取第2小特征值。
五、从二类别聚类到多类别聚类
1、二类别聚类
对于求解出来的特征向量中的每一个分量,根据每个分量的值来判断对应的点所属的类别:
2、多类别聚类
对于求出来的前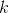 个特征向量,可以利用
K-Means
聚类方法对其进行聚类,若前
个特征向量,可以利用
K-Means
聚类方法对其进行聚类,若前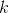 个特征向量为
个特征向量为 ,这样便由特征向量构成如下的特征向量矩阵:
,这样便由特征向量构成如下的特征向量矩阵:
将特征向量矩阵中的每一行最为一个样本,利用K-Means聚类方法对其进行聚类。
六、谱聚类的过程
1、基本的结构
基于以上的分析,谱聚类的基本过程为:
- 对于给定的图,求图的度矩阵
 和邻接矩阵
和邻接矩阵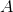 ;
; - 计算图的Laplacian矩阵;
- 对Laplacian矩阵进行特征值分解,取其前
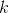 个特征值对应的特征向量,构成的特征向量矩阵;
个特征值对应的特征向量,构成的特征向量矩阵; - 利用K-Means聚类算法对上述的的特征向量矩阵进行聚类,每一行代表一个样本点。
2、利用相似度矩阵的构造方法
上述的方法是通过图的度矩阵 和邻接矩阵
和邻接矩阵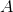 来构造Laplacian矩阵,也可以通过相似度矩阵的方法构造Laplacian矩阵,其方法如下:
来构造Laplacian矩阵,也可以通过相似度矩阵的方法构造Laplacian矩阵,其方法如下:
相似度矩阵是由权值矩阵得到:
其中
再利用相似度矩阵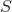 构造
Laplacian
矩阵:
构造
Laplacian
矩阵:
其中 为相似度矩阵
为相似度矩阵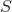 的度矩阵。
的度矩阵。
注意:在第一种方法中,求解的是Laplacian矩阵的前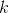 个最小特征值对应的特征向量,在第二种方法中,求解的是Laplacian矩阵的前
个最小特征值对应的特征向量,在第二种方法中,求解的是Laplacian矩阵的前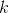 个最大特征值对应的特征向量
个最大特征值对应的特征向量
七、实验代码
1、自己实现的一个
#coding:UTF-8
'''
Created on 2015年5月12日
@author: zhaozhiyong
'''
from __future__ import division
import scipy.io as scio
from scipy import sparse
from scipy.sparse.linalg.eigen import arpack#这里只能这么做,不然始终找不到函数eigs
from numpy import *
def spectalCluster(data, sigma, num_clusters):
print "将邻接矩阵转换成相似矩阵"
#先完成sigma != 0
print "Fixed-sigma谱聚类"
data = sparse.csc_matrix.multiply(data, data)
data = -data / (2 * sigma * sigma)
S = sparse.csc_matrix.expm1(data) + sparse.csc_matrix.multiply(sparse.csc_matrix.sign(data), sparse.csc_matrix.sign(data))
#转换成Laplacian矩阵
print "将相似矩阵转换成Laplacian矩阵"
D = S.sum(1)#相似矩阵是对称矩阵
D = sqrt(1 / D)
n = len(D)
D = D.T
D = sparse.spdiags(D, 0, n, n)
L = D * S * D
#求特征值和特征向量
print "求特征值和特征向量"
vals, vecs = arpack.eigs(L, k=num_clusters,tol=0,which="LM")
# 利用k-Means
print "利用K-Means对特征向量聚类"
#对vecs做正规化
sq_sum = sqrt(multiply(vecs,vecs).sum(1))
m_1, m_2 = shape(vecs)
for i in xrange(m_1):
for j in xrange(m_2):
vecs[i,j] = vecs[i,j]/sq_sum[i]
myCentroids, clustAssing = kMeans(vecs, num_clusters)
for i in xrange(shape(clustAssing)[0]):
print clustAssing[i,0]
def randCent(dataSet, k):
n = shape(dataSet)[1]
centroids = mat(zeros((k,n)))#create centroid mat
for j in range(n):#create random cluster centers, within bounds of each dimension
minJ = min(dataSet[:,j])
rangeJ = float(max(dataSet[:,j]) - minJ)
centroids[:,j] = mat(minJ + rangeJ * random.rand(k,1))
return centroids
def distEclud(vecA, vecB):
return sqrt(sum(power(vecA - vecB, 2))) #la.norm(vecA-vecB)
def kMeans(dataSet, k):
m = shape(dataSet)[0]
clusterAssment = mat(zeros((m,2)))#create mat to assign data points to a centroid, also holds SE of each point
centroids = randCent(dataSet, k)
clusterChanged = True
while clusterChanged:
clusterChanged = False
for i in range(m):#for each data point assign it to the closest centroid
minDist = inf; minIndex = -1
for j in range(k):
distJI = distEclud(centroids[j,:],dataSet[i,:])
if distJI < minDist:
minDist = distJI; minIndex = j
if clusterAssment[i,0] != minIndex: clusterChanged = True
clusterAssment[i,:] = minIndex,minDist**2
#print centroids
for cent in range(k):#recalculate centroids
ptsInClust = dataSet[nonzero(clusterAssment[:,0].A==cent)[0]]#get all the point in this cluster
centroids[cent,:] = mean(ptsInClust, axis=0) #assign centroid to mean
return centroids, clusterAssment
if __name__ == '__main__':
# 导入数据集
matf = 'E://data_sc//corel_50_NN_sym_distance.mat'
dataDic = scio.loadmat(matf)
data = dataDic['A']
# 谱聚类的过程
spectalCluster(data, 20, 18)
2、网上提供的一个Matlab代码
function [cluster_labels evd_time kmeans_time total_time] = sc(A, sigma, num_clusters)
%SC Spectral clustering using a sparse similarity matrix (t-nearest-neighbor).
%
% Input : A : N-by-N sparse distance matrix, where
% N is the number of data
% sigma : sigma value used in computing similarity,
% if 0, apply self-tunning technique
% num_clusters : number of clusters
%
% Output : cluster_labels : N-by-1 vector containing cluster labels
% evd_time : running time for eigendecomposition
% kmeans_time : running time for k-means
% total_time : total running time
%
% Convert the sparse distance matrix to a sparse similarity matrix,
% where S = exp^(-(A^2 / 2*sigma^2)).
% Note: This step can be ignored if A is sparse similarity matrix.
%
disp('Converting distance matrix to similarity matrix...');
tic;
n = size(A, 1);
if (sigma == 0) % Selftuning spectral clustering
% Find the count of nonzero for each column
disp('Selftuning spectral clustering...');
col_count = sum(A~=0, 1)';
col_sum = sum(A, 1)';
col_mean = col_sum ./ col_count;
[x y val] = find(A);
A = sparse(x, y, -val.*val./col_mean(x)./col_mean(y)./2);
clear col_count col_sum col_mean x y val;
else % Fixed-sigma spectral clustering
disp('Fixed-sigma spectral clustering...');
A = A.*A;
A = -A/(2*sigma*sigma);
end
% Do exp function sequentially because of memory limitation
num = 2000;
num_iter = ceil(n/num);
S = sparse([]);
for i = 1:num_iter
start_index = 1 + (i-1)*num;
end_index = min(i*num, n);
S1 = spfun(@exp, A(:,start_index:end_index)); % sparse exponential func
S = [S S1];
clear S1;
end
clear A;
toc;
%
% Do laplacian, L = D^(-1/2) * S * D^(-1/2)
%
disp('Doing Laplacian...');
D = sum(S, 2) + (1e-10);
D = sqrt(1./D); % D^(-1/2)
D = spdiags(D, 0, n, n);
L = D * S * D;
clear D S;
time1 = toc;
%
% Do eigendecomposition, if L =
% D^(-1/2) * S * D(-1/2) : set 'LM' (Largest Magnitude), or
% I - D^(-1/2) * S * D(-1/2): set 'SM' (Smallest Magnitude).
%
disp('Performing eigendecomposition...');
OPTS.disp = 0;
[V, val] = eigs(L, num_clusters, 'LM', OPTS);
time2 = toc;
%
% Do k-means
%
disp('Performing kmeans...');
% Normalize each row to be of unit length
sq_sum = sqrt(sum(V.*V, 2)) + 1e-20;
U = V ./ repmat(sq_sum, 1, num_clusters);
clear sq_sum V;
cluster_labels = k_means(U, [], num_clusters);
total_time = toc;
%
% Calculate and show time statistics
%
evd_time = time2 - time1
kmeans_time = total_time - time2
total_time
disp('Finished!');
function cluster_labels = k_means(data, centers, num_clusters)
%K_MEANS Euclidean k-means clustering algorithm.
%
% Input : data : N-by-D data matrix, where N is the number of data,
% D is the number of dimensions
% centers : K-by-D matrix, where K is num_clusters, or
% 'random', random initialization, or
% [], empty matrix, orthogonal initialization
% num_clusters : Number of clusters
%
% Output : cluster_labels : N-by-1 vector of cluster assignment
%
% Reference: Dimitrios Zeimpekis, Efstratios Gallopoulos, 2006.
% http://scgroup.hpclab.ceid.upatras.gr/scgroup/Projects/TMG/
%
% Parameter setting
%
iter = 0;
qold = inf;
threshold = 0.001;
%
% Check if with initial centers
%
if strcmp(centers, 'random')
disp('Random initialization...');
centers = random_init(data, num_clusters);
elseif isempty(centers)
disp('Orthogonal initialization...');
centers = orth_init(data, num_clusters);
end
%
% Double type is required for sparse matrix multiply
%
data = double(data);
centers = double(centers);
%
% Calculate the distance (square) between data and centers
%
n = size(data, 1);
x = sum(data.*data, 2)';
X = x(ones(num_clusters, 1), :);
y = sum(centers.*centers, 2);
Y = y(:, ones(n, 1));
P = X + Y - 2*centers*data';
%
% Main program
%
while 1
iter = iter + 1;
% Find the closest cluster for each data point
[val, ind] = min(P, [], 1);
% Sum up data points within each cluster
P = sparse(ind, 1:n, 1, num_clusters, n);
centers = P*data;
% Size of each cluster, for cluster whose size is 0 we keep it empty
cluster_size = P*ones(n, 1);
% For empty clusters, initialize again
zero_cluster = find(cluster_size==0);
if length(zero_cluster) > 0
disp('Zero centroid. Initialize again...');
centers(zero_cluster, :)= random_init(data, length(zero_cluster));
cluster_size(zero_cluster) = 1;
end
% Update centers
centers = spdiags(1./cluster_size, 0, num_clusters, num_clusters)*centers;
% Update distance (square) to new centers
y = sum(centers.*centers, 2);
Y = y(:, ones(n, 1));
P = X + Y - 2*centers*data';
% Calculate objective function value
qnew = sum(sum(sparse(ind, 1:n, 1, size(P, 1), size(P, 2)).*P));
mesg = sprintf('Iteration %d:\n\tQold=%g\t\tQnew=%g', iter, full(qold), full(qnew));
disp(mesg);
% Check if objective function value is less than/equal to threshold
if threshold >= abs((qnew-qold)/qold)
mesg = sprintf('\nkmeans converged!');
disp(mesg);
break;
end
qold = qnew;
end
cluster_labels = ind';
%-----------------------------------------------------------------------------
function init_centers = random_init(data, num_clusters)
%RANDOM_INIT Initialize centroids choosing num_clusters rows of data at random
%
% Input : data : N-by-D data matrix, where N is the number of data,
% D is the number of dimensions
% num_clusters : Number of clusters
%
% Output: init_centers : K-by-D matrix, where K is num_clusters
rand('twister', sum(100*clock));
init_centers = data(ceil(size(data, 1)*rand(1, num_clusters)), :);
function init_centers = orth_init(data, num_clusters)
%ORTH_INIT Initialize orthogonal centers for k-means clustering algorithm.
%
% Input : data : N-by-D data matrix, where N is the number of data,
% D is the number of dimensions
% num_clusters : Number of clusters
%
% Output: init_centers : K-by-D matrix, where K is num_clusters
%
% Find the num_clusters centers which are orthogonal to each other
%
Uniq = unique(data, 'rows'); % Avoid duplicate centers
num = size(Uniq, 1);
first = ceil(rand(1)*num); % Randomly select the first center
init_centers = zeros(num_clusters, size(data, 2)); % Storage for centers
init_centers(1, :) = Uniq(first, :);
Uniq(first, :) = [];
c = zeros(num-1, 1); % Accumalated orthogonal values to existing centers for non-centers
% Find the rest num_clusters-1 centers
for j = 2:num_clusters
c = c + abs(Uniq*init_centers(j-1, :)');
[minimum, i] = min(c); % Select the most orthogonal one as next center
init_centers(j, :) = Uniq(i, :);
Uniq(i, :) = [];
c(i) = [];
end
clear c Uniq;
个人的一点认识:谱聚类的过程相当于先进行一个非线性的降维,然后在这样的低维空间中再利用聚类的方法进行聚类。
欢迎大家一起讨论,如有问题欢迎留言,欢迎大家转载。
参考
1、从拉普拉斯矩阵说到谱聚类(http://blog.csdn.net/v_july_v/article/details/40738211)
2、谱聚类(spectral clustering)(http://www.cnblogs.com/FengYan/archive/2012/06/21/2553999.html)
3、谱聚类算法(Spectral Clustering)(http://www.cnblogs.com/sparkwen/p/3155850.html)