刷题ROSALIND,练编程水平
http://rosalind.info/problems/list-view/
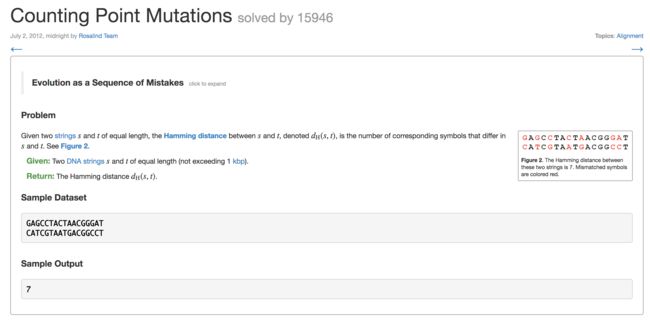
6. Counting Point Mutations(点突变计数)
- 解题思路
- 找两个序列的差异
s = 'GAGCCTACTAACGGGAT'
t = 'CATCGTAATGACGGCCT'
count = 0
for i in range(len(s)):
if s[i] != t[i]:
count +=1
print (count)
7
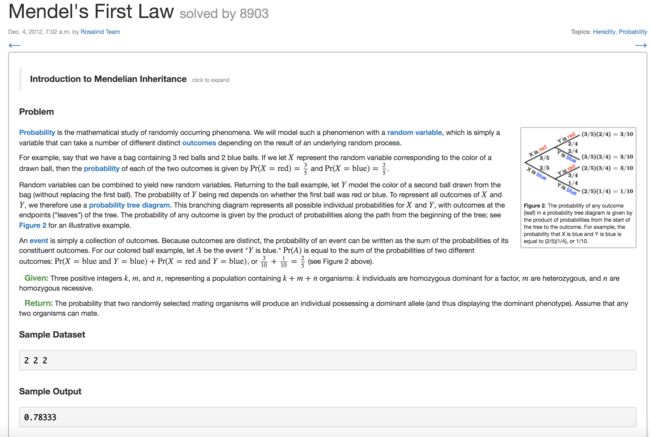
7. Mendel's First Law(孟德尔第一定律)
- 解题思路
-
有三个亲本(AA, Aa,aa)各两人,随机选择 两个亲本,1个后代个体表现显性性状的概率
-
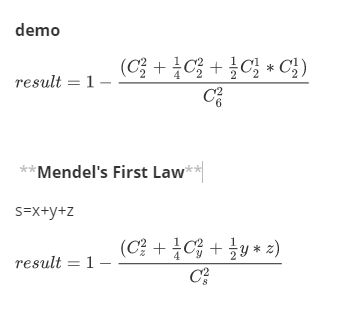
def f(x, y, z):
s = x + y + z # the sum of population
c = s * (s - 1) / 2.0 # comb(2,s)
# 1-(aa与aa+Aa与Aa+Aa与aa)
p = 1 - (z * (z - 1) / 2 + 0.25 * y * (y - 1) / 2 + y * z * 0.5) / c
return p
print (f(2, 2, 2))
0.7833333333333333
# -*- coding: utf-8 -*-
### 7. Mendel's First Law ###
from scipy.misc import comb
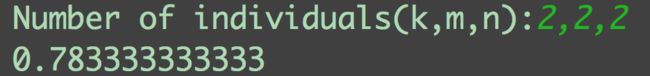
individuals = input('Number of individuals(k,m,n):')
# 把字符串映射成list
[k, m, n] = map(int, individuals.split(','))
t = k + m + n
rr = comb(n, 2) / comb(t, 2)
hh = comb(m, 2) / comb(t, 2)
hr = comb(n, 1) * comb(m, 1) / comb(t, 2)
prob = 1 - (rr + hh * 1 / 4 + hr * 1 / 2)
print(prob)
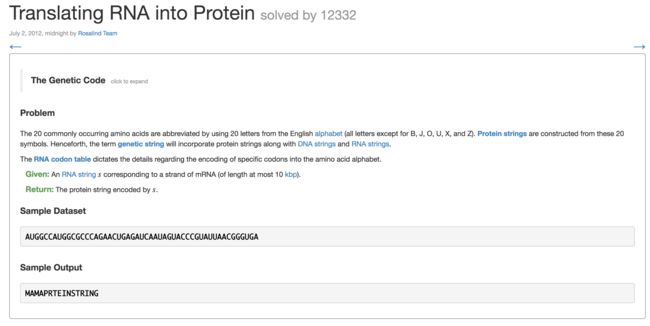
8. Translating RNA into Protein(RNA转蛋白质)
- 解题思路
- 需要氨基酸对应蛋白质的字典
- 匹配重写字符窜
def translate_rna(sequence):
# 密码子表
codonTable = {
'AUA': 'I', 'AUC': 'I', 'AUU': 'I', 'AUG': 'M',
'ACA': 'T', 'ACC': 'T', 'ACG': 'T', 'ACU': 'T',
'AAC': 'N', 'AAU': 'N', 'AAA': 'K', 'AAG': 'K',
'AGC': 'S', 'AGU': 'S', 'AGA': 'R', 'AGG': 'R',
'CUA': 'L', 'CUC': 'L', 'CUG': 'L', 'CUU': 'L',
'CCA': 'P', 'CCC': 'P', 'CCG': 'P', 'CCU': 'P',
'CAC': 'H', 'CAU': 'H', 'CAA': 'Q', 'CAG': 'Q',
'CGA': 'R', 'CGC': 'R', 'CGG': 'R', 'CGU': 'R',
'GUA': 'V', 'GUC': 'V', 'GUG': 'V', 'GUU': 'V',
'GCA': 'A', 'GCC': 'A', 'GCG': 'A', 'GCU': 'A',
'GAC': 'D', 'GAU': 'D', 'GAA': 'E', 'GAG': 'E',
'GGA': 'G', 'GGC': 'G', 'GGG': 'G', 'GGU': 'G',
'UCA': 'S', 'UCC': 'S', 'UCG': 'S', 'UCU': 'S',
'UUC': 'F', 'UUU': 'F', 'UUA': 'L', 'UUG': 'L',
'UAC': 'Y', 'UAU': 'Y', 'UAA': '', 'UAG': '',
'UGC': 'C', 'UGU': 'C', 'UGA': '', 'UGG': 'W',
}
proteinsequence = ''
# 3个3个取
for n in range(0, len(sequence), 3):
if sequence[n:n + 3] in codonTable.keys():
# 把匹配到的字典的键值加入到蛋白质字符窜
proteinsequence += codonTable[sequence[n:n + 3]]
return proteinsequence
se = "AUGGCCAUGGCGCCCAGAACUGAGAUCAAUAGUACCCGUAUUAACGGGUGA" # sequence
print(translate_rna(se))
MAMAPRTEINSTRING
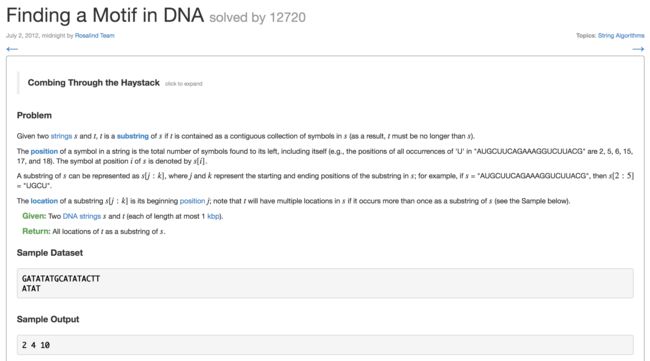
9. Finding a Motif in DNA(在DNA中找模体)
- 解题思路
- 在“GATATATGCATACTT” 中找ATAT,找出现的坐标,多次匹配多次出现的坐标
- 用正则表达式
import regex
# 如果overlapped 为False, 匹配到位置为2,10
matches = regex.finditer('ATAT', 'GATATATGCATATACTT', overlapped=True)
# 返回所有匹配项,
for match in matches:
print(match.start() + 1)
2 4 10
seq = 'GATATATGCATATACTT'
pattern = 'ATAT'
def find_motif(seq, pattern):
position = []
for i in range(len(seq) - len(pattern)):
if seq[i:i + len(pattern)] == pattern:
position.append(str(i + 1))
print('\t'.join(position))
find_motif(seq, pattern)
2 4 10
# methond 3
import re
seq = 'GATATATGCATATACTT'
# ?= 之后字符串内容需要匹配表达式才能成功匹配。
for i in re.finditer('(?=ATAT)', seq):
print(i.start() + 1)
2 4 10
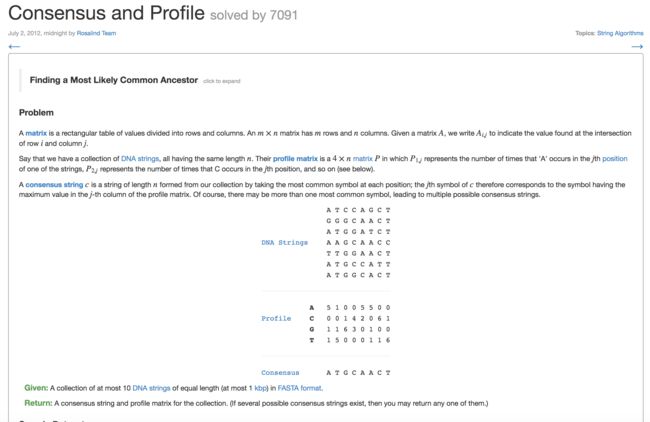
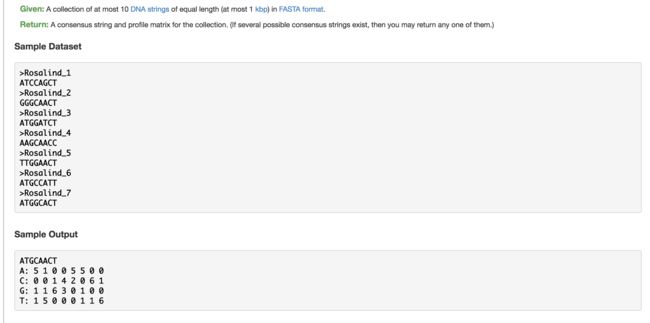
10. Consensus and Profile(寻找一致序列)
- 解题思路
- 共有序列:多条相同长度DNA字符串,对每个位置,选择出现最多的那个字符
# -*-coding:UTF-8-*-
### 10. Consensus and Profile ###
from collections import Counter
from collections import OrderedDict
# 写出到文件
fh = open('consesus_and_profile_output.txt', 'wt')
rosalind = OrderedDict()
seqLength = 0
with open('10 Consensus and Profile') as f:
for line in f:
line = line.rstrip()
if line.startswith('>'):
rosalindName = line
rosalind[rosalindName] = ''
# 跳出这个循环,进入下一个循环
continue
# 如果不是'>'开头,执行这一句
rosalind[rosalindName] += line
# len(ATCCAGCT)
seqLength = len(rosalind[rosalindName])
A, C, G, T = [], [], [], []
consensusSeq = ''
for i in range(seqLength):
seq = ''
# rosalind.keys = Rosalind_1...Rosalind_7
for k in rosalind.keys():
# 把 Rosalind_1...Rosalind_7 相同顺序的碱基加起来
seq += rosalind[k][i]
A.append(seq.count('A'))
C.append(seq.count('C'))
G.append(seq.count('G'))
T.append(seq.count('T'))
# 为seq计数
counts = Counter(seq)
# 从多到少返回,是个list啊,只返回第一个
consensusSeq += counts.most_common()[0][0]
fh.write(consensusSeq + '\n')
fh.write('\n'.join(['A:\t' + '\t'.join(map(str, A)), 'C:\t' + '\t'.join(map(str, C)),
'G:\t' + '\t'.join(map(str, G)), 'T:\t' + '\t'.join(map(str, T))]))
# .join(map(str,A)) 把 A=[5, 1, 0, 0, 5, 5, 0, 0] 格式转化成 A:5 1 0 0 5 5 0 0
fh.close()
# coding=utf-8
import numpy as np
import os
from collections import Counter
fhand = open('10 Consensus and Profile')
t = []
for line in fhand:
if line.startswith('>'):
continue
else:
line = line.rstrip()
line_list = list(line) # 变成一个list
t.append(line_list) # 再把list 放入list
a = np.array(t) # 创建一个二维数组
L1, L2, L3, L4 = [], [], [], []
comsquence = ''
for i in range(a.shape[1]): # shape[0],是7 行数,shape[1]是8 项目数
l = [x[i] for x in a] # 调出二维数组的每一列
L1.append(l.count('A'))
L2.append(l.count('C'))
L3.append(l.count('T'))
L4.append(l.count('G'))
comsquence = comsquence + Counter(l).most_common()[0][0]
print(comsquence)
print('A:', L1, '\n', 'C:', L2, '\n', 'T:', L3, '\n', 'G:', L4)
- 欢迎交流