scikit-learn 逻辑回归实现乳腺癌检测
随书代码,阅读笔记
- 载入数据
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
# 载入数据
from sklearn.datasets import load_breast_cancer
cancer = load_breast_cancer()
X = cancer.data
y = cancer.target
print('data shape: {0}; no. positive: {1}; no. negative: {2}'.format(
X.shape, y[y==1].shape[0], y[y==0].shape[0]))
print(cancer.data[0])
#准备测试集和训练集
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)一共有569个样本,每个样本有30个特征,其中357个阳性,212个阴性(y=0)
- 模型训练
# 模型训练
from sklearn.linear_model import LogisticRegression
model = LogisticRegression()
model.fit(X_train, y_train)
train_score = model.score(X_train, y_train)
test_score = model.score(X_test, y_test)
print('train score: {train_score:.6f}; test score: {test_score:.6f}'.format(
train_score=train_score, test_score=test_score))
#output: train score: 0.953846; test score: 0.956140- 预测
# 样本预测
y_pred = model.predict(X_test)
print('matchs: {0}/{1}'.format(np.equal(y_pred, y_test).shape[0], y_test.shape[0]))
# 预测概率:找出低于 90% 概率的样本个数
y_pred_proba = model.predict_proba(X_test)
print('sample of predict probability: {0}'.format(y_pred_proba[0]))
y_pred_proba_0 = y_pred_proba[:, 0] > 0.1
result = y_pred_proba[y_pred_proba_0]
y_pred_proba_1 = result[:, 1] > 0.1
print(result[y_pred_proba_1])模型优化
import time
from sklearn.linear_model import LogisticRegression
from sklearn.preprocessing import PolynomialFeatures
from sklearn.pipeline import Pipeline
# 增加多项式预处理
def polynomial_model(degree=1, **kwarg):
polynomial_features = PolynomialFeatures(degree=degree,
include_bias=False)
logistic_regression = LogisticRegression(**kwarg)
pipeline = Pipeline([("polynomial_features", polynomial_features),
("logistic_regression", logistic_regression)])
return pipeline
model = polynomial_model(degree=2, penalty='l1')
start = time.clock()
model.fit(X_train, y_train)
train_score = model.score(X_train, y_train)
cv_score = model.score(X_test, y_test)
print('elaspe: {0:.6f}; train_score: {1:0.6f}; cv_score: {2:.6f}'.format(
time.clock()-start, train_score, cv_score))
#output : train_score: 1.000000; cv_score: 0.973684新特征
根据原始的30个特征,使用多项式组合出来495个特征,其中97个是有用的。
logistic_regression = model.named_steps['logistic_regression']
print('model parameters shape: {0}; count of non-zero element: {1}'.format(
logistic_regression.coef_.shape,
np.count_nonzero(logistic_regression.coef_)))
#output:model parameters shape: (1, 495); count of non-zero element: 97学习率曲线
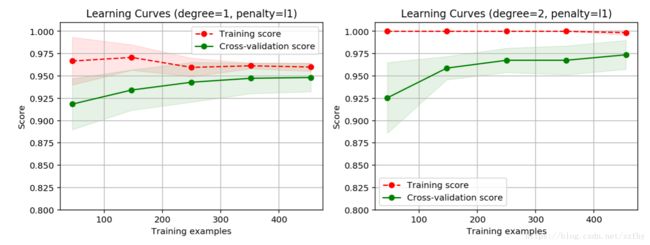
from common.utils import plot_learning_curve
from sklearn.model_selection import ShuffleSplit
cv = ShuffleSplit(n_splits=10, test_size=0.2, random_state=0)
title = 'Learning Curves (degree={0}, penalty={1})'
degrees = [1, 2]
penalty = 'l1'
start = time.clock()
plt.figure(figsize=(12, 4), dpi=144)
for i in range(len(degrees)):
plt.subplot(1, len(degrees), i + 1)
plot_learning_curve(plt, polynomial_model(degree=degrees[i], penalty=penalty),
title.format(degrees[i], penalty), X, y, ylim=(0.8, 1.01), cv=cv)
print('elaspe: {0:.6f}'.format(time.clock()-start))
penalty = 'l2'
start = time.clock()
plt.figure(figsize=(12, 4), dpi=144)
for i in range(len(degrees)):
plt.subplot(1, len(degrees), i + 1)
plot_learning_curve(plt, polynomial_model(degree=degrees[i], penalty=penalty, solver='lbfgs'),
title.format(degrees[i], penalty), X, y, ylim=(0.8, 1.01), cv=cv)
print('elaspe: {0:.6f}'.format(time.clock()-start))扩展阅读