Matplotlib画图包与实例
文章目录
- Bar Plot
- Scatter Matrix Plot
- 3D Scatter Plot
- Decision Boundary Plot
- Scatter Plot
-
- (I) K-NN classfication accuracy versus sensitivity to K
- (II) K-NN classfication accuracy versus sensitivity to train-test proportion
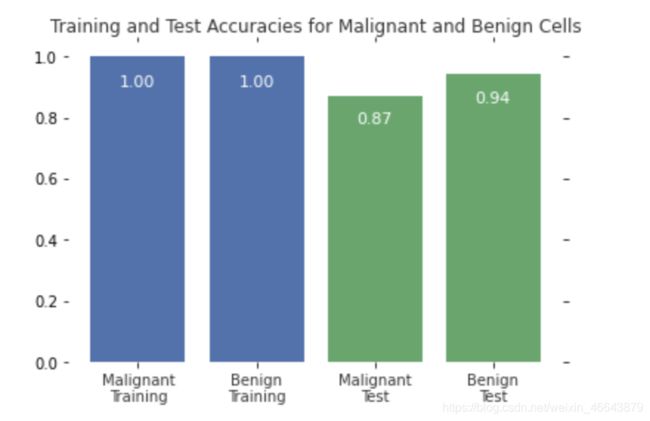
Bar Plot
- X轴label
- Bar颜色选择和数据标记
- Title
- 去框线
import matplotlib.pyplot as plt
plt.figure()
bars=plt.bar(np.arange(4),scores,color=['#4c72b0','#4c72b0','#55a868','#55a868'])
#label score onto the bar
for bar in bars:
height=bar.get_height()
plt.gca().text(bar.get_x()+ bar.get_width()/2,height*.90,'{0:.{1}f}'.format(height, 2),ha='center', color='w', fontsize=11)
#remove all the ticks (both axes), and tick labels on the Y axis
plt.tick_params(top='off', bottom='off', left='off', right='off', labelleft='off', labelbottom='on')
#remove the frame of the chart
for spine in plt.gca().spines.values():
spine.set_visible(False)
plt.xticks([0,1,2,3], ['Malignant\nTraining', 'Benign\nTraining', 'Malignant\nTest', 'Benign\nTest'], alpha=0.8);
plt.title('Training and Test Accuracies for Malignant and Benign Cells', alpha=0.8)
plt.figure()
plt.bar()
plt.gca().text()
plt.gca().spines.values()
spine.set_visible()
plt.xticks()
plt.title()
bar.get_height()
bar.get_x()
bar.get_width()
Scatter Matrix Plot
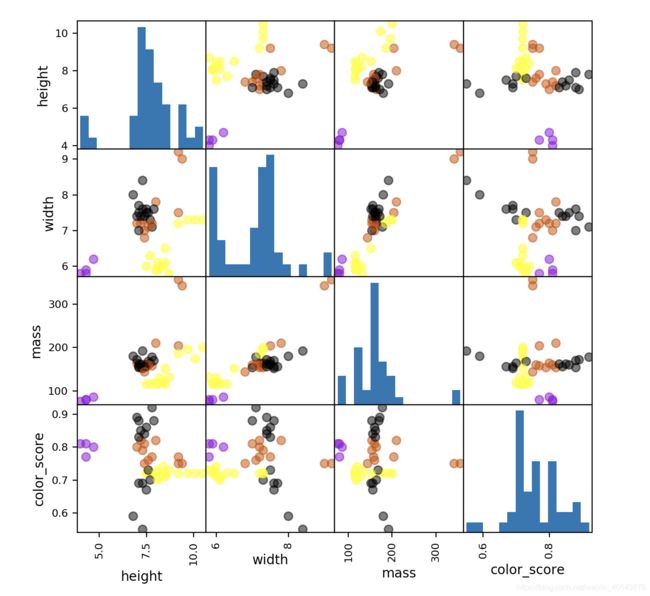
Data consists of 4 features + one label
# plotting a scatter matrix
from matplotlib import cm #color map
X = fruits[['height', 'width', 'mass', 'color_score']]
y = fruits['fruit_label']
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
cmap = cm.get_cmap('gnuplot')
scatter = pd.scatter_matrix(X_train, c= y_train, marker = 'o', s=40, hist_kwds={
'bins':15}, figsize=(7,7), cmap=cmap)
cm.get_cmap()
pd.scatter_matrix()
3D Scatter Plot
# plotting a 3D scatter plot
from mpl_toolkits.mplot3d import Axes3D
fig = plt.figure()
ax = fig.add_subplot(111, projection = '3d')
ax.scatter(X_train['width'], X_train['height'], X_train['color_score'], c = y_train, marker = 'o', s=100)
ax.set_xlabel('width')
ax.set_ylabel('height')
ax.set_zlabel('color_score')
plt.show()
fig=plt.figure()
ax=fig.add_subplot()
ax.scatter()
ax.set_xlabel()
ax.set_ylabel()
ax.set_zlabel()
plt.show()
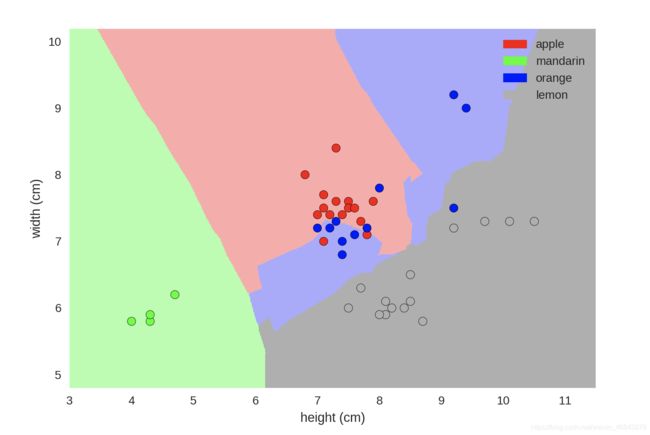
Decision Boundary Plot
from adspy_shared_utilities import plot_fruit_knn
plot_fruit_knn(X_train, y_train, 5, 'uniform') # we choose 5 nearest neighbors
Scatter Plot
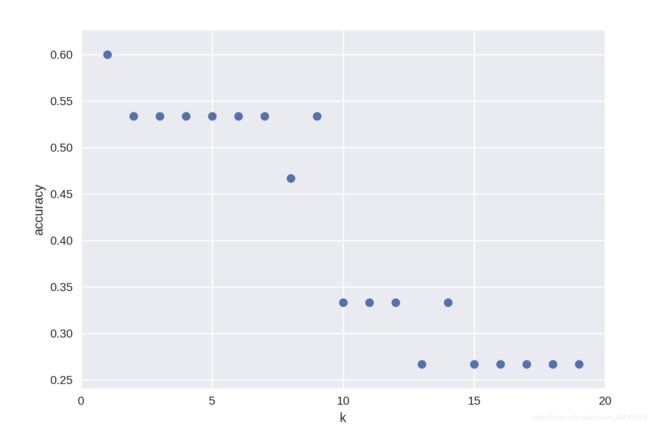
(I) K-NN classfication accuracy versus sensitivity to K
import matplotlib.pyplot as plt
k_range = range(1,20)
scores = []
for k in k_range:
knn = KNeighborsClassifier(n_neighbors = k)
knn.fit(X_train, y_train)
scores.append(knn.score(X_test, y_test))
plt.figure()
plt.xlabel('k')
plt.ylabel('accuracy')
plt.scatter(k_range, scores)
plt.xticks([0,5,10,15,20]);
plt.figure()
plt.scatter()
plt.xlabel()
plt.ylabel()
plt.xticks()
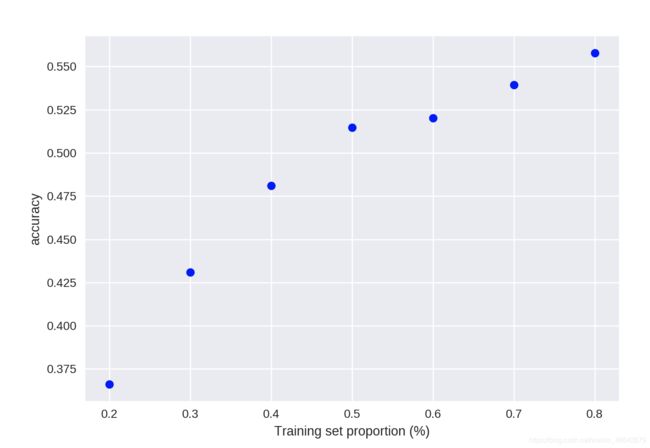
(II) K-NN classfication accuracy versus sensitivity to train-test proportion
import matplotlib.pyplot as plt
t = [0.8, 0.7, 0.6, 0.5, 0.4, 0.3, 0.2]
knn = KNeighborsClassifier(n_neighbors = 5)
plt.figure()
for s in t:
scores = []
for i in range(1,1000):
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 1-s)
knn.fit(X_train, y_train)
scores.append(knn.score(X_test, y_test))
plt.plot(s, np.mean(scores), 'bo')
plt.xlabel('Training set proportion (%)')
plt.ylabel('accuracy');
plt.figure()
plt.plot()
plt.xlabel()
plt.ylabel()