化合物分子 ogb、dgl生成图网络及GNN模型训练;pgl图框架
参考:https://towardsdatascience.com/learn-to-smell-molecules-with-graph-convolutional-neural-networks-62fa5a826af5
https://github.com/snap-stanford/ogb
https://github.com/dmlc/dgl/blob/master/examples/pytorch
https://programtalk.com/python-more-examples/ogb.utils.features.atom_to_feature_vector/
https://keras.io/examples/generative/wgan-graphs/
ogb 包是一个图数据操作、加载
1、ogb、dgl simles生成图网络
import torch
import dgl
import torch_geometric
from ogb.utils.features import atom_to_feature_vector, bond_to_feature_vector, get_atom_feature_dims, \
get_bond_feature_dims
from rdkit import Chem
from rdkit.Chem.rdmolops import GetAdjacencyMatrix
from torch.utils.data import Dataset
import numpy as np
import pandas as pd
from tqdm import tqdm
import torch.nn.functional as F
from scipy.constants import physical_constants
from typing import List, Tuple
def graph_only_collate(batch: List[Tuple]):
return dgl.batch(batch)
class InferenceDataset(Dataset):
def __init__(self, smiles_txt_path, device='cuda:0', transform=None, **kwargs):
with open(smiles_txt_path) as file:
lines = file.readlines()
smiles_list = [line.rstrip() for line in lines]
atom_slices = [0]
edge_slices = [0]
all_atom_features = []
all_edge_features = []
edge_indices = [] # edges of each molecule in coo format
total_atoms = 0
total_edges = 0
n_atoms_list = []
for mol_idx, smiles in tqdm(enumerate(smiles_list)):
# get the molecule using the smiles representation from the csv file
mol = Chem.MolFromSmiles(smiles)
# add hydrogen bonds to molecule because they are not in the smiles representation
mol = Chem.AddHs(mol)
n_atoms = mol.GetNumAtoms()
atom_features_list = []
for atom in mol.GetAtoms():
atom_features_list.append(atom_to_feature_vector(atom))
all_atom_features.append(torch.tensor(atom_features_list, dtype=torch.long))
edges_list = []
edge_features_list = []
for bond in mol.GetBonds():
i = bond.GetBeginAtomIdx()
j = bond.GetEndAtomIdx()
edge_feature = bond_to_feature_vector(bond)
# add edges in both directions
edges_list.append((i, j))
edge_features_list.append(edge_feature)
edges_list.append((j, i))
edge_features_list.append(edge_feature)
# Graph connectivity in COO format with shape [2, num_edges]
edge_index = torch.tensor(edges_list, dtype=torch.long).T
edge_features = torch.tensor(edge_features_list, dtype=torch.long)
edge_indices.append(edge_index)
all_edge_features.append(edge_features)
total_edges += len(edges_list)
total_atoms += n_atoms
edge_slices.append(total_edges)
atom_slices.append(total_atoms)
n_atoms_list.append(n_atoms)
self.n_atoms = torch.tensor(n_atoms_list)
self.atom_slices = torch.tensor(atom_slices, dtype=torch.long)
self.edge_slices = torch.tensor(edge_slices, dtype=torch.long)
self.edge_indices = torch.cat(edge_indices, dim=1)
self.all_atom_features = torch.cat(all_atom_features, dim=0)
self.all_edge_features = torch.cat(all_edge_features, dim=0)
def __len__(self):
return len(self.atom_slices) - 1
def __getitem__(self, idx):
e_start = self.edge_slices[idx]
e_end = self.edge_slices[idx + 1]
start = self.atom_slices[idx]
n_atoms = self.n_atoms[idx]
edge_indices = self.edge_indices[:, e_start: e_end]
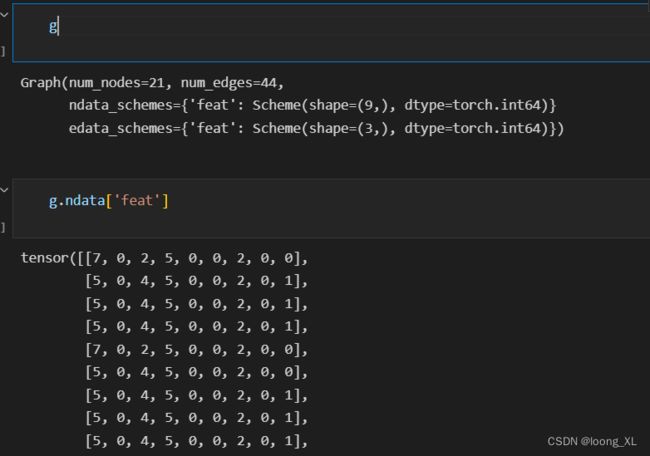
g = dgl.graph((edge_indices[0], edge_indices[1]), num_nodes=n_atoms)
g.ndata['feat'] = self.all_atom_features[start: start + n_atoms]
g.edata['feat'] = self.all_edge_features[e_start: e_end]
return g
test_data = InferenceDataset(device=device, smiles_txt_path=args.smiles_txt_path) ## simles 字符串txt,每行一个
test_loader = DataLoader(test_data, batch_size=2, collate_fn=graph_only_collate) ## torch的DataLoader
单个测试
smiles_list =["OC1CC1(O)CC1CC1"]
atom_slices = [0]
edge_slices = [0]
all_atom_features = []
all_edge_features = []
edge_indices = [] # edges of each molecule in coo format
total_atoms = 0
total_edges = 0
n_atoms_list = []
for mol_idx, smiles in tqdm(enumerate(smiles_list)):
# get the molecule using the smiles representation from the csv file
mol = Chem.MolFromSmiles(smiles)
# add hydrogen bonds to molecule because they are not in the smiles representation
mol = Chem.AddHs(mol)
print(Chem.MolToSmiles(mol))
n_atoms = mol.GetNumAtoms()
print(n_atoms)
atom_features_list = []
for atom in mol.GetAtoms():
atom_features_list.append(atom_to_feature_vector(atom))
all_atom_features.append(torch.tensor(atom_features_list, dtype=torch.long))
edges_list = []
edge_features_list = []
for bond in mol.GetBonds():
i = bond.GetBeginAtomIdx()
j = bond.GetEndAtomIdx()
edge_feature = bond_to_feature_vector(bond)
# add edges in both directions
edges_list.append((i, j))
edge_features_list.append(edge_feature)
edges_list.append((j, i))
edge_features_list.append(edge_feature)
# Graph connectivity in COO format with shape [2, num_edges]
edge_index = torch.tensor(edges_list, dtype=torch.long).T
edge_features = torch.tensor(edge_features_list, dtype=torch.long)
edge_indices.append(edge_index)
all_edge_features.append(edge_features)
total_edges += len(edges_list)
total_atoms += n_atoms
edge_slices.append(total_edges)
atom_slices.append(total_atoms)
n_atoms_list.append(n_atoms)
n_atoms = torch.tensor(n_atoms_list)
atom_slices = torch.tensor(atom_slices, dtype=torch.long)
edge_slices = torch.tensor(edge_slices, dtype=torch.long)
edge_indices = torch.cat(edge_indices, dim=1)
all_atom_features = torch.cat(all_atom_features, dim=0)
all_edge_features = torch.cat(all_edge_features, dim=0)
e_start = edge_slices[0]
e_end = edge_slices[0 + 1]
start = atom_slices[0]
n_atoms = n_atoms[0]
edge_indices = edge_indices[:, e_start: e_end]
g = dgl.graph((edge_indices[0], edge_indices[1]), num_nodes=n_atoms)
g.ndata['feat'] = all_atom_features[start: start + n_atoms]
g.edata['feat'] = all_edge_features[e_start: e_end]
2、GNN模型训练代码
row.csv
SMILES,SENTENCE
C/C=C/C(=O)C1CCC(C=C1C)(C)C,"fruity,rose"
COC(=O)OC,"fresh,ethereal,fruity"
Cc1cc2c([nH]1)cccc2,"resinous,animalic"
C1CCCCCCCC(=O)CCCCCCC1,"powdery,musk,animalic"
CC(CC(=O)OC1CC2C(C1(C)CC2)(C)C)C,"coniferous,camphor,fruity"
CCC[C@H](CCO)SC,tropicalfruit
from rdkit import Chem
import numpy as np
import pandas as pd
from tqdm import tqdm
import torch
import dgl
from ogb.utils.features import atom_to_feature_vector, bond_to_feature_vector, get_atom_feature_dims, \
get_bond_feature_dims
def smiles2graph(smiles_string):
mol = Chem.MolFromSmiles(smiles_string)
A = Chem.GetAdjacencyMatrix(mol)
A = np.asmatrix(A)
nz = np.nonzero(A)
src, dst = nz[0], nz[1]
g = dgl.graph((src, dst))
return g
def feat_vec(smiles_string):
"""
Returns atom features for a molecule given a smiles string
"""
# atoms
mol = Chem.MolFromSmiles(smiles_string)
atom_features_list = []
for atom in mol.GetAtoms():
atom_features_list.append(atom_to_feature_vector(atom))
x = np.array(atom_features_list, dtype = np.int64)
return x
df =pd.read_csv("row.csv")
lista_senten=df['SENTENCE'].to_list()
labels=[]
for olor in lista_senten:
olor=olor.split(",")
if 'fruity' in olor:
labels.append(1)
else:
labels.append(0)
lista_mols=df['SMILES'].to_list()
j=0
graphs=[]
execptions=[]
for mol in lista_mols:
g_mol=smiles2graph(mol)
try:
g_mol.ndata['feat']=torch.tensor(feat_vec(mol))
except:
execptions.append(j)
graphs.append(g_mol)
j+=1
from dgl.data import DGLDataset
class SyntheticDataset(DGLDataset):
def __init__(self):
super().__init__(name='synthetic')
def process(self):
self.graphs = graphs
self.labels = torch.LongTensor(labels)
def __getitem__(self, i):
return self.graphs[i], self.labels[i]
def __len__(self):
return len(self.graphs)
dataset = SyntheticDataset()
from dgl.dataloading import GraphDataLoader
from torch.utils.data.sampler import SubsetRandomSampler
num_examples = len(dataset)
num_train = int(num_examples * 0.8)
train_sampler = SubsetRandomSampler(torch.arange(num_train))
test_sampler = SubsetRandomSampler(torch.arange(num_train, num_examples))
train_dataloader = GraphDataLoader(
dataset, sampler=train_sampler, batch_size=5, drop_last=False)
test_dataloader = GraphDataLoader(
dataset, sampler=test_sampler, batch_size=5, drop_last=False)
from dgl.nn import GraphConv
from torch import nn
import torch.nn.functional as F
class GCN(nn.Module):
def __init__(self, in_feats, h_feats, num_classes):
super(GCN, self).__init__()
self.conv1 = GraphConv(in_feats, h_feats)
self.conv2 = GraphConv(h_feats, num_classes)
def forward(self, g, in_feat):
h = self.conv1(g, in_feat)
h = F.relu(h)
h = self.conv2(g, h)
g.ndata['h'] = h
return dgl.mean_nodes(g, 'h')
model = GCN(9, 8, 2)
optimizer = torch.optim.Adam(model.parameters(), lr=0.01)
for epoch in range(20):
for batched_graph, labels in train_dataloader:
pred = model(batched_graph, batched_graph.ndata['feat'].float())
#print(pred,labels)
loss = F.cross_entropy(pred, labels)
optimizer.zero_grad()
loss.backward()
optimizer.step()
num_correct = 0
num_tests = 0
for batched_graph, labels in test_dataloader:
pred = model(batched_graph, batched_graph.ndata['feat'].float())
num_correct += (pred.argmax(1) == labels).sum().item()
num_tests += len(labels)
print('Test accuracy:', num_correct / num_tests)
##保存
torch.save(
model.state_dict(), os.path.join(args.save_dir, args.name)
)
## 先加载模型框架
model.load_state_dict(torch.load(os.path.join(args.save_dir, args.name)))
2、pgl图框架安装
参考:https://pytorch-geometric.readthedocs.io/en/latest/notes/installation.html
这款是torch原生;上面dgl是亚马逊先开发的也比较通用
pgl容易出错,建议conda 创建个python==3.9环境,然后pgl对于的一些列包如果安装报Microsoft Visual C++ 14.0 or greater is required. Get it wit等错误,建议直接下载wheel轮子进行安装
https://pytorch-geometric.com/whl wheel轮子地址;这里安装的torch对应torch=1.12.1,windows上安装
pip install torch-scatter torch-sparse torch-cluster torch-spline-conv torch-geometric