RDKit|摩根分子指纹计算、提取与可视化
文章目录
- 一、摩根分子指纹计算
- 1.简介
- 2.SparseIntVects
- 3.ExplicitBitVects
- 4.FCFPs
- 5.更多泛化
- 二、摩根分子指纹提取
- 1.提取方法一
- 2.提取方法二
- 三、指纹可视化
一、摩根分子指纹计算
1.简介
摩根分子指纹(Morgan Fingerprints),是一种圆形指纹,也属于拓扑型指纹,是通过对标准的摩根算法进行改造后得到。可以大致等同于扩展连通性指纹(Extended-Connectivity Fingerprints,ECFPs)。
这类指纹有诸多优点,例如计算速度快、没有经过预定义(可以表示无穷多种不同的分子特征)、可以包含手性信息、指纹中的每个元素代表一种特定子结构、可以方便地进行分析和解释、可以根据不同的需要进行相应的修改等。这类指纹设计的最初目的是用于搜索与活性相关的分子特征,而非子结构搜索。此外也可以用于相似性搜索、聚类、虚拟筛选等方向。指纹的生成过程大致分为以下几个步骤:
- 1.原子初始化。为每个重原子分配一个整数标识符
- 2.迭代更新。以每个重原子为中心,将周围一圈的重原子合并进来,直到到达指定半径。
- 3.特征生成。对子结构进行运算,并生成特征列表。
更多内容可以参考ChemAxon的介绍,还有ECFPs的文章。
ECFPs可以捕捉到精确的子结构细节,相对应的,功能基指纹(Functional class fingerprints,FCFPs)则更为泛化,可以将同一类功能基作为一种特征结构。在rdkit中两种特征都可以通过GetMorganFingerprint实现。
- 先定义两个分子
>>> from rdkit import Chem
>>> from rdkit import DataStructs
>>> from rdkit.Chem import AllChem
>>> from rdkit.Chem import Draw
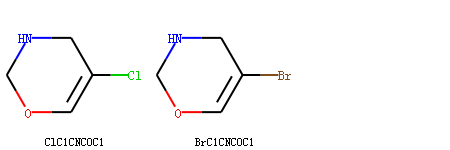
>>> m1 = Chem.MolFromSmiles('ClC1=COCNC1')
>>> m2 = Chem.MolFromSmiles('BrC1=COCNC1')
>>> Draw.MolsToGridImage([m1, m2], subImgSize=(150, 150), legends=['ClC1CNCOC1', 'BrC1CNCOC1'])
2.SparseIntVects
- 以SparseIntVects方式生成ECFPs指纹:GetMorganFingerprint(mol, radius, …)
或者称为SparseBitVects
mol:mol对象
radius:考虑半径 - 使用Dice方法比较两个指纹的相似性:DiceSimilarity()
>>> fp1 = AllChem.GetMorganFingerprint(m1, 2)
>>> fp2 = AllChem.GetMorganFingerprint(m2, 2)
>>> DataStructs.DiceSimilarity(fp1, fp2)
0.7
- 查看长度:GetLength()
返回指纹的长度 - 查看非空元素:GetNonzeroElements()
返回值是一个字典,键对应非空指纹的位数,值是指纹出现的频数
>>> print(fp1.GetLength())
4294967295
>>> print(fp1.GetNonzeroElements())
{39328034: 1, 211414882: 1, 362715007: 1, 2626911012: 1, 2968968094: 2, ...}
3.ExplicitBitVects
- 以ExplicitBitVects方式生成ECFPs指纹:GetMorganFingerprintAsBitVect(mol, radius, nBits, …)
radius:考虑半径
nBits:长度
>>> fp1 = AllChem.GetMorganFingerprintAsBitVect(m1, 2, nBits=1024)
>>> fp2 = AllChem.GetMorganFingerprintAsBitVect(m2, 2, nBits=1024)
>>> DataStructs.DiceSimilarity(fp1, fp2)
0.6842105263157895
4.FCFPs
- 通过设置useFeatures=True生成FCFPs
可以看到ffp1和ffp2完全一致,即Cl和Br在特征上被归为一类
>>> ffp1 = AllChem.GetMorganFingerprintAsBitVect(m1, 2, nBits=10, useFeatures=True)
>>> ffp2 = AllChem.GetMorganFingerprintAsBitVect(m2, 2, nBits=10, useFeatures=True)
>>> DataStructs.DiceSimilarity(ffp1, ffp2)
1.0
- 查看向量长度:GetNumBits()
- 将指纹转成字符串:ToBitString()
>>> print(ffp1.GetNumBits())
10
>>> print(ffp1.ToBitString())
1111101111
5.更多泛化
- 通过设置invariants,忽略原子类型,关注分子骨架
先来初始化三个分子
>>> m2 = Chem.MolFromSmiles('BrC1=CCCCC1')
>>> m3 = Chem.MolFromSmiles('BrC1CCCCC1')
>>> Draw.MolsToGridImage([m1, m2, m3], subImgSize=(200, 150))
>>> fp1 = AllChem.GetMorganFingerprint(m1, 2, invariants=[1]*m1.GetNumAtoms())
>>> fp2 = AllChem.GetMorganFingerprint(m2, 2, invariants=[1]*m2.GetNumAtoms())
>>> fp3 = AllChem.GetMorganFingerprint(m3, 2, invariants=[1]*m3.GetNumAtoms())
>>> fp1 == fp2
True
- 比较fp1和fp2,可以看到指纹忽略了N、O、C等差异,即fp1和fp2指纹一致,但这个时候还会考虑键的信息。例如比较fp1和fp3,它们是不相等的。
>>> fp1 == fp3
False
- 通过设置useBondTypes,忽略键的类型
>>> fp1 = AllChem.GetMorganFingerprint(m1, 2, invariants=[1]*m1.GetNumAtoms(), useBondTypes=False)
>>> fp3 = AllChem.GetMorganFingerprint(m1, 2, invariants=[1]*m3.GetNumAtoms(), useBondTypes=False)
>>> fp1 == fp3
True
二、摩根分子指纹提取
- 在生成分子指纹过程中,通过向参数bitInfo传入字典,可以获取所有非空指纹信息
字典info键表示位的索引,值为原子索引、半径构成的元组,出现多个元组时,表示子结构出现了多次
键为2968968094的值中含有多个元组,表示原子索引为4,6,半径为0的子结构相同,都记录在这一位上
>>> info = {}
>>> fp_explain = AllChem.GetMorganFingerprint(m1, 2, bitInfo=info)
>>> info
{39328034: ((1, 1),),
211414882: ((5, 2),),
362715007: ((6, 1),),
397705891: ((4, 1),),
718785834: ((1, 2),),
1016841875: ((0, 0),),
1078999752: ((3, 1),),
1289643292: ((5, 1),),
2132511834: ((5, 0),),
2626911012: ((4, 2),),
2968968094: ((4, 0), (6, 0)),
...}
1.提取方法一
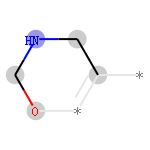
- 以提取出211414882的结构为例,首先提取m1中,半径为2,第5个原子的结构
submol接收提取出的子结构
amap用于接收原子索引的映射关系,键为原始分子中的原子索引,值为子结构中的原子索引
env是被提取出的键的索引
>>> amap = {}
>>> env = Chem.FindAtomEnvironmentOfRadiusN(m1, 2, 5)
>>> submol=Chem.PathToSubmol(m1, env, atomMap=amap)
>>> print(amap)
{1: 4, 3: 0, 4: 1, 5: 2, 6: 3}
>>> print(list(env))
[4, 5, 3, 6]
- 用SMILES表示该子结构
>>> Chem.MolToSmiles(submol)
'CCNCO'
- 以第5个原子为中心,用SMILES表示该子结构,可以看出是氮原子
>>> Chem.MolToSmiles(submol, rootedAtAtom=amap[5], canonical=False)
'N(CO)CC'
2.提取方法二
- 遍历env中的所有键,获取键连接的所有原子,保存在atoms中
通过给定分子,及要提取的原子信息、键信息,获取子结构
>>> atoms=set()
>>> for bidx in env:
>>> atoms.add(m1.GetBondWithIdx(bidx).GetBeginAtomIdx())
>>> atoms.add(m1.GetBondWithIdx(bidx).GetEndAtomIdx())
>>> Chem.MolFragmentToSmiles(m1, atomsToUse=list(atoms), bondsToUse=env, rootedAtAtom=5)
'N(CC)CO'
三、指纹可视化
查看211414882代表的子结构
- 绘制子结构:DrawMorganBit(mol, bitId, bitInfo, …)
mol:mol对象
bitId:要查看的bit id
bitInfo:指纹信息
>>> Draw.DrawMorganBit(m1, 211414882, info)
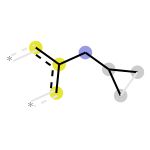
- 在ECFPs中的颜色的含义:
蓝色:表示中心原子
黄色:表示带有芳香性质的原子
灰色:表示在脂肪环中的原子
>>> mol = Chem.MolFromSmiles('c1ccccc1CC1CC1')
>>> bi = {}
>>> fp = AllChem.GetMorganFingerprintAsBitVect(mol, radius=2, bitInfo=bi)
>>> Draw.DrawMorganBit(mol, 872, bi)