本文是Johns Hopkins Univerisity
文章基于Rmarkdown编写的(需要安装Rstudio和knitr)
重点集成学习算法:baggig,random forest, boosting
predicting with tree基于树的机器学习
basic algorithm
1.start with all variables in one group
2.find the varialbe/split thata best separates the outcomes
3.divide the data into two groups on that split
4.within each split,find the best variable/split that the outcomes
5.continue until the groups are too small or sufficiently “pure”
measures of impurity
1.misclassification error
2.gini index
3.deviance/information gain
data(iris)
library(ggplot2)
names(iris)
## [1] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width"
## [5] "Species"
library(caret)
table(iris$Species)
## ## setosa versicolor virginica
## 50 50 50
creat train and test set
intrainiris <- createDataPartition(y=iris$Species,p=0.7,list=FALSE)
trainingiris <-iris[intrainiris,]
testingiris <- iris[-intrainiris,]
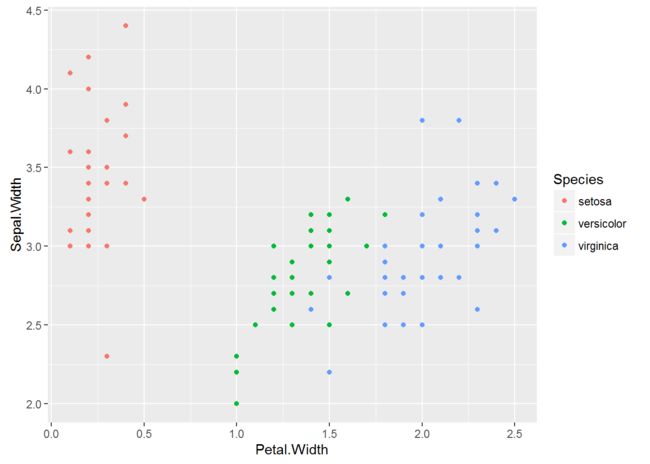
plot petal widths/sepal width
ggplot(trainingiris,aes(Petal.Width,Sepal.Width,color=Species))+geom_point()
model
modelfitiris<-train(Species~.,method="rpart",data=trainingiris)
print(modelfitiris$finalModel)
## n= 105
##
## node), split, n, loss, yval, (yprob)
## * denotes terminal node
##
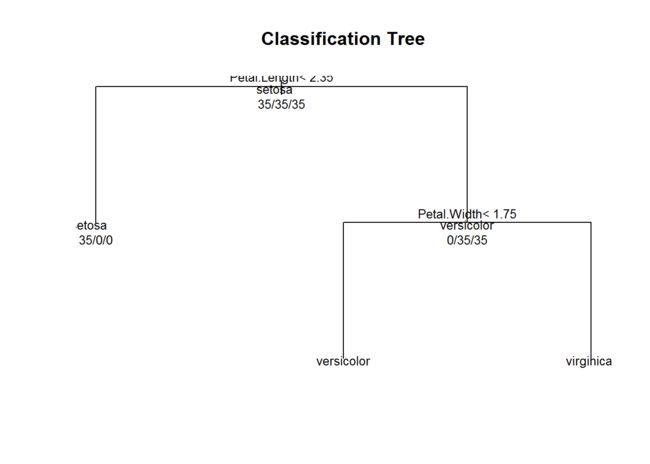
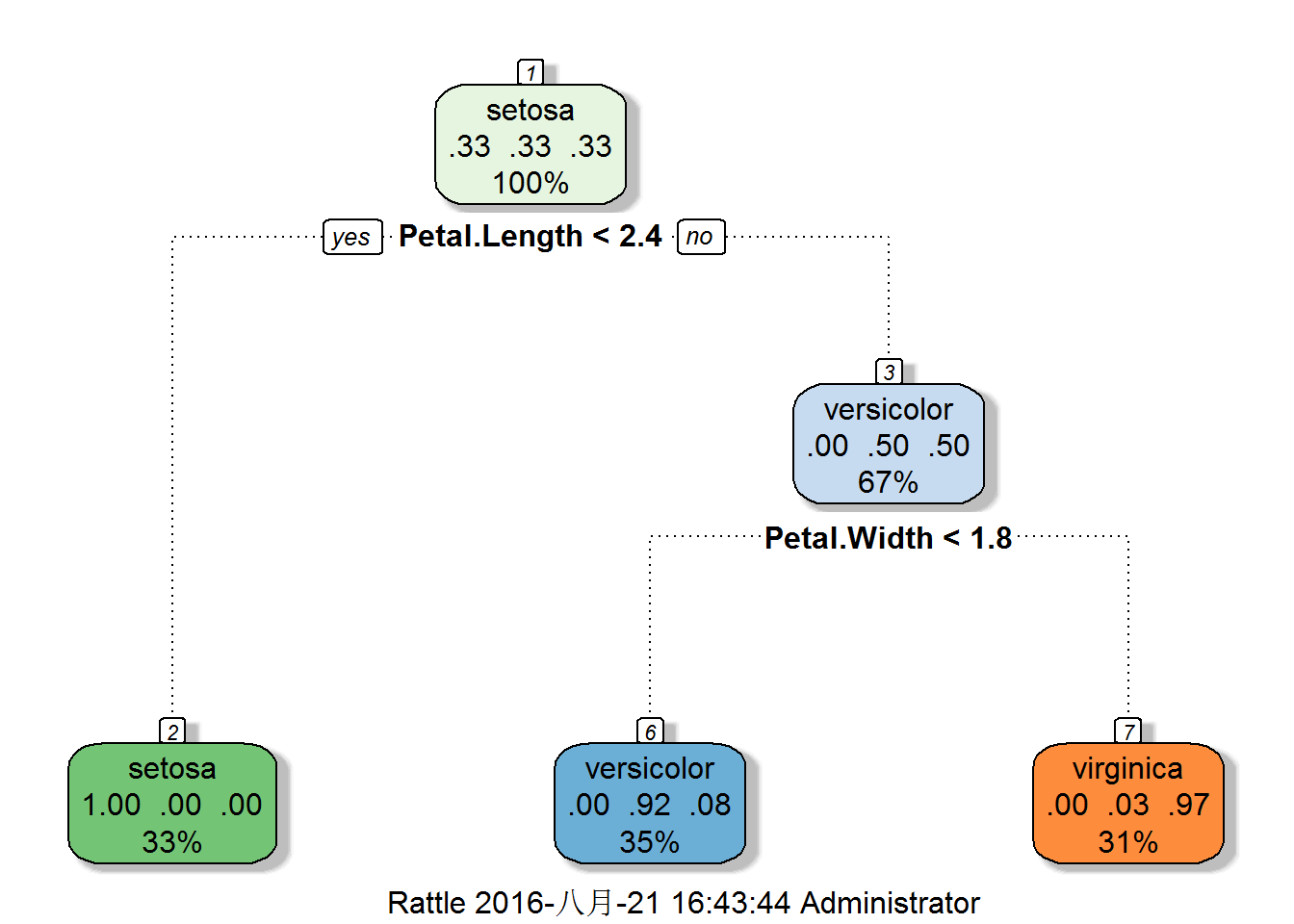
## 1) root 105 70 setosa (0.33333333 0.33333333 0.33333333)
## 2) Petal.Length< 2.35 35 0 setosa (1.00000000 0.00000000 0.00000000) *
## 3) Petal.Length>=2.35 70 35 versicolor (0.00000000 0.50000000 0.50000000)
## 6) Petal.Width< 1.75 37 3 versicolor (0.00000000 0.91891892 0.08108108) *
## 7) Petal.Width>=1.75 33 1 virginica (0.00000000 0.03030303 0.96969697) *
plot tree
plot(modelfitiris$finalModel,uniform = TRUE,main="Classification Tree")
text(modelfitiris$finalModel,use.n=TRUE,all=TRUE,cex=0.8)
Prettier plots
#install.packages("rattle")
#install.packages("rpart.plot")
library(rattle)library(rpart)
fancyRpartPlot(modelfitiris$finalModel)
predicting new values
predict(modelfitiris,newdata = testingiris)
## [1] setosa setosa setosa setosa setosa setosa
## [7] setosa setosa setosa setosa setosa setosa
## [13] setosa setosa setosa versicolor versicolor versicolor
## [19] versicolor versicolor versicolor versicolor versicolor versicolor
## [25] versicolor versicolor versicolor versicolor versicolor versicolor
## [31] virginica versicolor virginica virginica virginica virginica
## [37] virginica virginica versicolor virginica virginica virginica
## [43] virginica virginica virginica
## Levels: setosa versicolor virginica
modelevalation
TBD~~~~
bagging
resample cases and recalculate predictions
average or majority vote
notes:similar
bias;
reduced variance;
more useful for non-linear functions
import Ozone data
#install.packages("ElemStatLearn")
library(ElemStatLearn)
data(ozone,package = "ElemStatLearn")
ozone <-ozone[order(ozone$ozone),]
head(ozone)
## ozone radiation temperature wind
## 17 1 8 59 9.7
## 19 4 25 61 9.7
## 14 6 78 57 18.4
## 45 7 48 80 14.3
## 106 7 49 69 10.3
## 7 8 19 61 20.1
bagged loess?????
ll <-matrix(NA,nrow =10,ncol=155)
for (i in 1:10){
ss <- sample(1:dim(ozone)[1],replace=T)
ozone0 <- ozone[ss,]
ozone0<-ozone0[order(ozone0$ozone),]
loess0<-loess(temperature~ozone,data=ozone0,span=0.2)
ll[i,]<-predict(loess0,newdata=data.frame(ozone=1:155))}
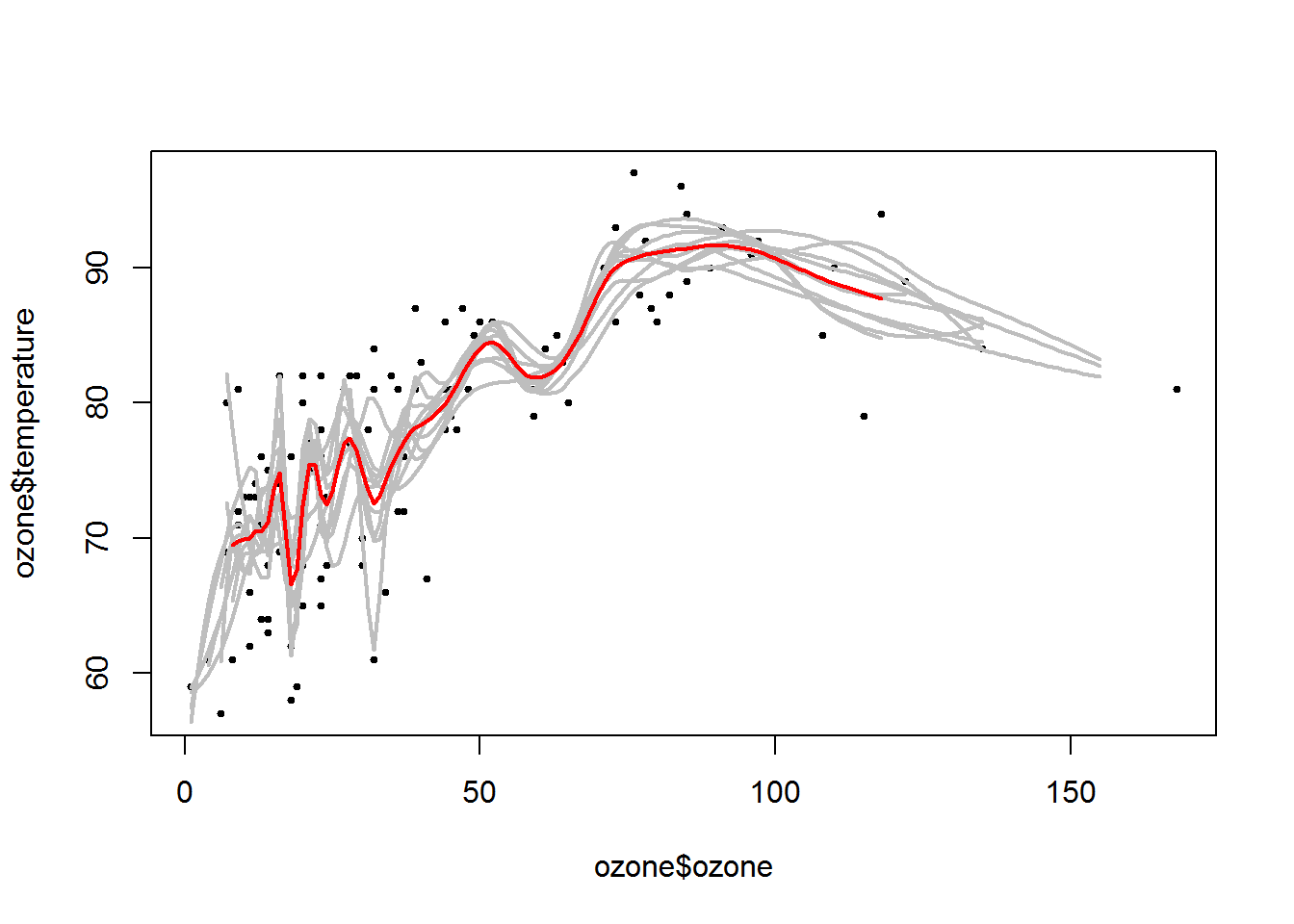
plot
plot(ozone$ozone,ozone$temperature,pch =19,cex=0.5)
for(i in 1:10){lines(1:155,ll[i,],col="grey",lwd=2)}
lines(1:155,apply(ll,2,mean),col="red",lwd=2)
set your own bagging定制bagging
predictiors =data.frame(ozone=ozone$ozone)
temperature=ozone$temperature
treebag<-bag(predictors,temperature,B=10,
bagControl=bagControl(fit=ctreeBag$fit,
predict=ctreeBag$pred,aggregate=ctreeBag$aggregate))
random Forests
bootstrap samples;
at each split,bootstrap variables;
grow multiple trees and vote
pros
accuracy
cons
speed;
interpretability;
overfitting(using CV)
model
modelfitrf <-train(Species~.,data=trainingiris,method="rf",prox=TRUE)
modelfitrf
## Random Forest
##
## 105 samples
## 4 predictor
## 3 classes: 'setosa', 'versicolor', 'virginica'
##
## No pre-processing
## Resampling: Bootstrapped (25 reps)
## Summary of sample sizes: 105, 105, 105, 105, 105, 105, ...
## Resampling results across tuning parameters:
##
## mtry Accuracy Kappa
## 2 0.9432666 0.9135607
## 3 0.9465044 0.9185112
## 4 0.9475875 0.9201665
##
## Accuracy was used to select the optimal model using the largest value.
## The final value used for the model was mtry = 4.
predicting new values
predirisrf <- predict(modelfitrf,testingiris)
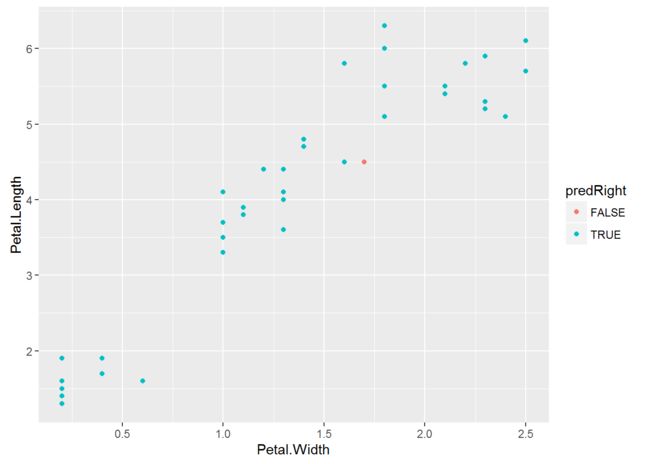
testingiris$predRight <-predirisrf==testingiris$Species
table(predirisrf,testingiris$Species)
##
## predirisrf setosa versicolor virginica
## setosa 15 0 0
## versicolor 0 15 1
## virginica 0 0 14
#plot pred TRUE and FALSE
ggplot(testingiris,aes(Petal.Width,Petal.Length,color=predRight,main=
"newdata Predictions"))+geom_point()
Boosting
basic idea
1.take lots of weak predictors
2.weight them and add them up
3.get a stronger predictor#boosting on R boosting
gbm boosting with trees
mboost model based boosting
ada statistical boosting base on additive logistic regression
gamBoost for boosting generalized additive modelsmost of them are available on caret package
library(ISLR)
data(Wage)
Wage <- subset(Wage,select=-c(logwage))
intrainwage<- createDataPartition(y=Wage$wage,p=0.7,list =FALSE)#return matrix
trainingwage<- Wage[intrainwage,]
testingwage<-Wage[-intrainwage,]
model
modelfitgbm <-train(wage~.,method="gbm",data=trainingwage,verbose=FALSE)
print(modelfitgbm)
## Stochastic Gradient Boosting
##
## 2102 samples
## 10 predictor
##
## No pre-processing
## Resampling: Bootstrapped (25 reps)
## Summary of sample sizes: 2102, 2102, 2102, 2102, 2102, 2102, ...
## Resampling results across tuning parameters:
##
## interaction.depth n.trees RMSE Rsquared
## 1 50 33.96349 0.3087931
## 1 100 33.43872 0.3209031
## 1 150 33.40399 0.3225494
## 2 50 33.42910 0.3217876
## 2 100 33.37711 0.3240077
## 2 150 33.44096 0.3228785
## 3 50 33.39148 0.3227759
## 3 100 33.51229 0.3204942
## 3 150 33.68149 0.3158711
##
## Tuning parameter 'shrinkage' was held constant at a value of 0.1
##
## Tuning parameter 'n.minobsinnode' was held constant at a value of 10
## RMSE was used to select the optimal model using the smallest value.
## The final values used for the model were n.trees = 100,
## interaction.depth = 2, shrinkage = 0.1 and n.minobsinnode = 10.
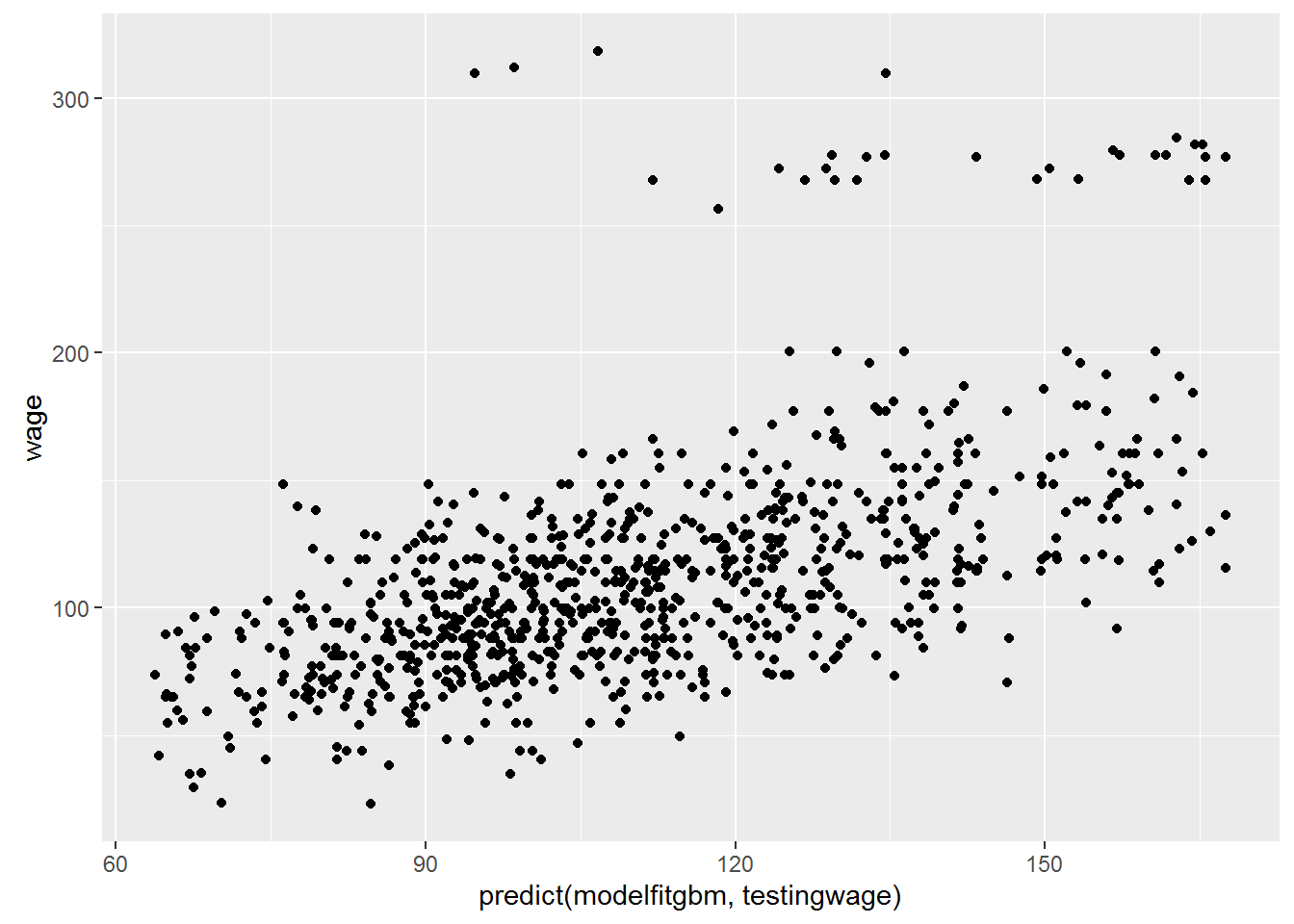
ggplot(testingwage,aes(predict(modelfitgbm,testingwage),wage))+geom_point()
model based prediction
linear discriminant analysis
naive bayes
modelfitlda<-train(Species~.,data=trainingiris,method="lda")
modelfitnb <-train(Species~.,data=trainingiris,method="nb")
plda=predict(modelfitlda,testingiris)
pnb <-predict(modelfitnb,testingiris)table(plda,pnb)
## pnb
## plda setosa versicolor virginica
## setosa 15 0 0
## versicolor 0 15 0
## virginica 0 1 14