【论文笔记】HARP: Hierarchical Representation Learning for Networks
目录
Abstract
Introduction
问题定义
Method
源码【code】
[paper]https://arxiv.org/pdf/1706.07845v2.pdf
[code]https://github.com/GTmac/HARP
Abstract
Our proposed method achieves this by compressing the input graph prior to embedding it, effectively avoiding troublesome embedding configurations (i.e.local minima) which can pose problems to non-convex optimization.
HARP works by finding a smaller graph which approximates the global structure of its input. This simplified graph is used to learn a set of initial representations, which serve as good initializations for learning representations in the original, detailed graph. We inductively extend this idea, by decomposing a graph in a series of levels, and then embed the hierarchy of graphs from the coarsest one to the original graph.
-
论文提出一种在获取图结构embedding之前先进行压缩的操作,因为直接使用deepwalk、node2vec、line,对于大的图来说,可能只能获得一个局部的embedding,他们所捕捉的网络结点关系过近,都是局部邻居。LINE仅仅只考虑到了一阶邻居与二阶邻居之间的相似度关系。DeepWalk与Node2Vec虽然可以通过随机游走,获得较长的游走序列,但是游走的长度比起现在图数据的规模,随机游走的长度还是太短了。所以就需要想一种方法捕捉全局的相似度。
-
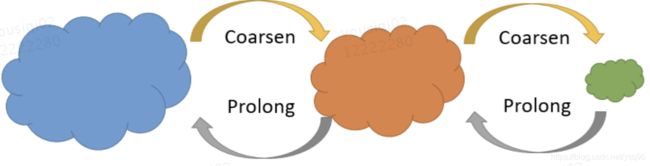
HARP采取的方式就是通过多次折叠,将原来的大图层层收缩为较小的图,使得通过较短的随机游走距离,就能够覆盖所有的网络结点,然后将小图作为DeepWalk、LINE或Node2Vec的输入,学习Embedding。最后将在收缩后的小图中学习到的Embedding作为折叠前的的图的Embedding的初始化,继续学习折叠前的图的Embedding。以此类推层层学得原来大图的Embedding。
Introduction
• New Representation Learning Paradigm. We proposeHARP, a novel multilevel paradigm for graph representation which seamlessly blends ideas from the graph drawing (Fruchterman and Reingold 1991) and graph representation learning (Perozzi, Al-Rfou, and Skiena 2014;Tang et al. 2015; Grover and Leskovec 2016) communities to build substantially better graph embeddings.
• Improved Optimization Primitives. We demonstrate that our approach leads to improved implementations of all state-of-the-art graph representation learning methods, namely DeepWalk (DW), LINE and Node2vec (N2V). Our improvements on these popular methods for learning latent representations illustrate the broad applicability of our hierarchical approach.
• Better Embeddings for Downstream Tasks. We demonstrate that HARP(DW), HARP(LINE) and HARP(N2V)embeddings consistently outperform the originals on classification tasks on several real-world networks, with improvements as large as 14% Macro F1.
-
主要的三点贡献:1⃣️一种新的网络表示学习例子2⃣️优化了原始的方法(deepwalk、node2vec、line)3⃣️为下游任务获得更好的embedding
问题定义
Method
-
HARP分为三步:1⃣️图折叠2⃣️图嵌入3⃣️表示的恢复
-
HARP的关键就是图折叠算法
-
边折叠:边折叠算法选择尽可能多的边,这些边没有共同的顶点,即每个顶点只有一条与之连接的边被选中。
-
星折叠 :虽然边折叠在最好的情况下每一轮折叠可以将结点的数量缩减一半,这样一来图折叠算法的总次数为O(logk)。但在某些特殊情况下却不能很好的发挥作用。如下图(b)所示,星形结构是网络中最常见结构之一,在这种结构中,如果使用边折叠算法,那么就至多只能折叠一条边,算法要执行的总次数则会退化为O(k)。所以,HARP采用了另外一种折叠策略,就是星折叠。
源码【code】
源码使用的是python2 gensim=0.13.2 scipy=0.19.1 容易由于和numpy、pandas版本发生冲突,这里做了修改
-
harp.py(执行函数)
import magicgraph import logging import os import sys import numpy as np from argparse import ArgumentParser, FileType, ArgumentDefaultsHelpFormatter from magicgraph import WeightedDiGraph, WeightedNode from scipy.io import mmread, mmwrite, loadmat import graph_coarsening def main(): parser = ArgumentParser('harp', formatter_class=ArgumentDefaultsHelpFormatter, conflict_handler='resolve') parser.add_argument('--format', default='mat', help='File format of input file') parser.add_argument('--input', nargs='?', required=True, help='Input graph file') parser.add_argument('--sfdp-path', default='./bin/sfdp_osx', help='Path to the SFDP binary file which produces graph coarsening results.') parser.add_argument('--model', default='deepwalk', help='Embedding model to use. Could be deepwalk, line or node2vec.') parser.add_argument('--matfile-variable-name', default='network', help='Variable name of adjacency matrix inside a .mat file') parser.add_argument('--number-walks', default=40, type=int, help='Number of random walks to start at each node') parser.add_argument('--output', required=True, help='Output representation file') parser.add_argument('--representation-size', default=128, type=int, help='Number of latent dimensions to learn for each node.') parser.add_argument('--walk-length', default=10, type=int, help='Length of the random walk started at each node.') parser.add_argument('--window-size', default=10, type=int, help='Window size of the Skip-gram model.') parser.add_argument('--workers', default=1, type=int, help='Number of parallel processes.') args = parser.parse_args() # Process args if args.format == 'mat': G = magicgraph.load_matfile(args.input, variable_name=args.matfile_variable_name, undirected=True) elif args.format == 'adjlist': G = magicgraph.load_adjacencylist(args.input, undirected=True) elif args.format == 'edgelist': # 读取边表 得到无向图 G = magicgraph.load_edgelist(args.input, undirected=True) else: raise Exception("Unknown file format: '%s'. Valid formats: 'mat', 'adjlist', and 'edgelist'." % args.format) G = graph_coarsening.DoubleWeightedDiGraph(G) print ('Number of nodes: {}'.format(G.number_of_nodes())) print ('Number of edges: {}'.format(G.number_of_edges())) print ('Underlying network embedding model: {}'.format(args.model)) # 主要差别在deepwalk采用 Skip-gram + Hierarchical Softmax # 其他方法采用的是 Skip-gram + Negative Sampling # 通过hs控制 if args.model == 'deepwalk': embeddings = graph_coarsening.skipgram_coarsening_disconnected(G,scale=-1,iter_count=1, sfdp_path=args.sfdp_path, num_paths=args.number_walks,path_length=args.walk_length, representation_size=args.representation_size,window_size=args.window_size, lr_scheme='default',alpha=0.025,min_alpha=0.001,sg=1,hs=1,coarsening_scheme=2, sample=0.1) elif args.model == 'node2vec': embeddings = graph_coarsening.skipgram_coarsening_disconnected(G,scale=-1,iter_count=1, sfdp_path=args.sfdp_path, num_paths=args.number_walks,path_length=args.walk_length, representation_size=args.representation_size,window_size=args.window_size, lr_scheme='default',alpha=0.025,min_alpha=0.001,sg=1,hs=0,coarsening_scheme=2, sample=0.1) elif args.model == 'line': embeddings = graph_coarsening.skipgram_coarsening_disconnected(G,scale=1, iter_count=50, sfdp_path=args.sfdp_path, representation_size=64,window_size=1, lr_scheme='default',alpha=0.025,min_alpha=0.001,sg=1,hs=0,sample=0.001) np.save(args.output, embeddings) if __name__ == '__main__': sys.exit(main()) -
graph_coarsening.py(图折叠的主要函数)
import copy import glob import logging import magicgraph import math import operator import os import random import skipgram import subprocess import sys import tempfile import baseline import utils import numpy as np from collections import defaultdict, deque from concurrent.futures import ProcessPoolExecutor from deepwalk import walks as serialized_walks from gensim.models import Word2Vec from magicgraph import WeightedDiGraph, WeightedNode from scipy.io import mmread, mmwrite class DoubleWeightedDiGraph(WeightedDiGraph): def __init__(self, init_graph = None): super(WeightedDiGraph, self).__init__(node_class=WeightedNode) self.weighted_nodes = magicgraph.WeightedNode() if init_graph is not None: for node, adj_list in init_graph.adjacency_iter(): if hasattr(adj_list, 'weights'): self[node].extend(adj_list, adj_list.weights) else: self[node].extend(adj_list, [1. for adj_node in adj_list]) if hasattr(init_graph, 'weighted_nodes'): self.weighted_nodes.extend(init_graph.nodes(), init_graph.weighted_nodes.weights) else: self.weighted_nodes.extend(init_graph.nodes(), [1. for node in init_graph.nodes()]) self.visited = {node: False for node in self.nodes()} def is_connected(self): # sys.setrecursionlimit(self.number_of_nodes()) self.visited = {node: False for node in self.nodes()} if self.number_of_nodes() == 0: return True self.cur_component = [] self.bfs(list(self.nodes())[0]) return sum(self.visited.values()) == self.number_of_nodes() def get_connected_components(self): connected_components = [] self.visited = {node: False for node in self.nodes()} for node in self.nodes(): if self.visited[node] is False: self.cur_component = [] self.bfs(node) connected_components.append(len(self.cur_component)) return connected_components # graph coarsening need to be done on each connected component def get_merged_connected_components(self): disconnected_component, connected_components, reversed_mappings = [], [], [] self.visited = {node: False for node in self.nodes()} graph_size_threshold = 100 for node in self.nodes(): if self.visited[node] is False: self.cur_component = [] self.bfs(node) if len(self.cur_component) >= graph_size_threshold: self.cur_component = sorted(self.cur_component) index_mapping = {self.cur_component[i]: i for i in range(len(self.cur_component)) } connected_components.append(self.subgraph(self.cur_component, index_mapping=index_mapping)) reversed_mappings.append({i: self.cur_component[i] for i in range(len(self.cur_component)) }) else: disconnected_component.extend(self.cur_component) if len(disconnected_component) > 0: disconnected_component = sorted(disconnected_component) reversed_mappings.append({i: disconnected_component[i] for i in range(len(disconnected_component)) }) index_mapping = {disconnected_component[i]: i for i in range(len(disconnected_component)) } connected_components.append(self.subgraph(disconnected_component, index_mapping=index_mapping) ) return connected_components, reversed_mappings def dfs(self, cur_node): self.visited[cur_node] = True self.cur_component.append(cur_node) for adj_node in self[cur_node]: if self.visited[adj_node] is False: self.visited[adj_node] = True self.dfs(adj_node) def bfs(self, cur_node): q = deque() q.append(cur_node) self.visited[cur_node] = True while len(q) > 0: head = q.popleft() self.cur_component.append(head) for adj_node in self[head]: if not self.visited[adj_node]: self.visited[adj_node] = True q.append(adj_node) def subgraph(self, nodes = {}, index_mapping = None): nodes = set(nodes) if index_mapping is None: index_mapping = {node: node for node in nodes} sub = DoubleWeightedDiGraph(magicgraph.from_adjlist([ [index_mapping[node]] for node in nodes])) for node in nodes: for adj_node, weight in zip(self[node], self[node].weights): if adj_node in nodes: sub[index_mapping[node]].append(index_mapping[adj_node], weight) if len(self[node]) == 0: if index_mapping: sub[index_mapping[node]].append(index_mapping[node], 1.) else: sub[node].append(node, 1.) node_weight_map = {node: weight for node, weight in zip(self.weighted_nodes, self.weighted_nodes.weights)} for node in nodes: sub.weighted_nodes.weights[index_mapping[node] ] = node_weight_map[node] return sub # get edges as pairs of integers def get_int_edges(self): edges, weights = [], [] for node in self.nodes(): for adj_node, weight in zip(self[node], self[node].weights): edges.append([node, adj_node]) weights.append(weight) return edges, weights # get edges along with weights def get_edges(self): edges, weights = [], [] for node in self.nodes(): for adj_node, weight in zip(self[node], self[node].weights): edges.append([str(node), str(adj_node)]) weights.append(weight) return edges, np.array(weights) def random_walk(self, path_length, alpha=0, rand=random.Random(), start=None): G = self if start is not None: path = [start] else: path = [rand.choice(G.keys())] while len(path) < path_length: cur = path[-1] if len(G[cur]) > 0: if rand.random() >= alpha: path.append(G[cur].choice(rand)) else: path.append(path[0]) else: break return path def external_collapsing(graph, merged): coarsened_graph = DoubleWeightedDiGraph() edges, weights = graph.get_int_edges() merged_edge_to_weight = defaultdict(float) node_weight = {node: weight for node, weight in zip(graph.weighted_nodes, graph.weighted_nodes.weights)} new_node_weights = defaultdict(float) for (a, b), w in zip(edges, weights): merged_a, merged_b = merged[a], merged[b] if merged_a != merged_b: merged_edge_to_weight[(merged_a, merged_b)] += w for node_pair, weight in merged_edge_to_weight.items(): coarsened_graph[node_pair[0]].append(node_pair[1], weight) coarsened_graph[node_pair[1]].append(node_pair[0], weight) for node in coarsened_graph.nodes(): coarsened_graph.weighted_nodes.append(node, new_node_weights[node]) return coarsened_graph.make_consistent() def read_coarsening_info(coarsening_file_dir): coarsening_files = [f for dirpath, dirnames, files in os.walk(coarsening_file_dir) for f in files if f.startswith('prolongation')] levels = -1 recursive_merged_nodes = [] for f in coarsening_files: levels = max(levels, int(f[f.rfind('_') + 1:]) ) prev_rename, rename = {}, {} for level in range(levels + 1): # different index merged_from = defaultdict(list) merged = {} fp = open(os.path.normpath(coarsening_file_dir) + '/' + 'prolongation_' + str(level)) for line in fp: finer_node, coarser_node = map(int, line.strip().split()) # let index starts from 0 instead finer_node, coarser_node = finer_node - 1, coarser_node - 1 if finer_node in prev_rename: # print coarser_node, finer_node, prev_rename[finer_node] merged_from[coarser_node].append(prev_rename[finer_node]) else: merged_from[coarser_node].append(finer_node) # print merged_from for k in merged_from.keys(): rename[k] = merged_from[k][0] for node in merged_from[k]: merged[node] = merged_from[k][0] # print merged recursive_merged_nodes.append(merged) prev_rename = rename.copy() rename = {} return recursive_merged_nodes def external_ec_coarsening(graph, sfdp_path, coarsening_scheme=2): temp_dir = tempfile.mkdtemp() temp_fname = 'tmp.mtx' input_fname = os.path.join(temp_dir, temp_fname) mmwrite(open(os.path.join(input_fname), 'wb'), magicgraph.to_adjacency_matrix(graph)) sfdp_abs_path = os.path.abspath(sfdp_path) subprocess.call('%s -g%d -v -u -Tc %s 2>x' % (sfdp_abs_path, coarsening_scheme, input_fname), shell=True, cwd=temp_dir) recursive_graphs, recursive_merged_nodes = [], read_coarsening_info(temp_dir) subprocess.call(['rm', '-r', temp_dir]) cur_graph = graph iter_round = 1 prev_node_count = graph.number_of_nodes() ec_done = False levels = len(recursive_merged_nodes) if levels == 0: return [graph], recursive_merged_nodes for level in range(levels): if iter_round == 1: print ('Original graph with %d nodes and %d edges' % \ (cur_graph.number_of_nodes(), cur_graph.number_of_edges() ) ) recursive_graphs.append(DoubleWeightedDiGraph(cur_graph)) coarsened_graph = external_collapsing(cur_graph, recursive_merged_nodes[level]) cur_node_count = coarsened_graph.number_of_nodes() print ('Coarsening Round %d:' % iter_round) print ('Generate coarsened graph with %d nodes and %d edges' % \ (coarsened_graph.number_of_nodes(), coarsened_graph.number_of_edges()) ) recursive_graphs.append(coarsened_graph) cur_graph = coarsened_graph iter_round += 1 prev_node_count = cur_node_count return recursive_graphs, recursive_merged_nodes def skipgram_coarsening_disconnected(graph, recursive_graphs=None, recursive_merged_nodes=None, **kwargs): # 输出参数 print (kwargs) # 图是否是连通图 if graph.is_connected(): print ('Connected graph.') # 若是连通图,则已经不能再折叠了,subgraphs即为graph subgraphs, reversed_mappings = [graph], [{node: node for node in graph.nodes()}] else: # 若不是连通图,则获取其subgraphs subgraphs, reversed_mappings = graph.get_merged_connected_components() count = 0 scale = kwargs.get('scale', -1) num_paths = kwargs.get('num_paths', 40) path_length = kwargs.get('path_length', 10) representation_size = kwargs.get('representation_size', 128) window_size = kwargs.get('window_size', 10) iter_count = kwargs.get('iter_count', 1) lr_scheme = kwargs.get('lr_scheme', 'default') alpha = kwargs.get('alpha', 0.025) min_alpha = kwargs.get('min_alpha', 0.001) report_loss = kwargs.get('report_loss', False) hs = kwargs.get('hs', 0) sample = kwargs.get('sample', 1e-3) coarsening_scheme = kwargs.get('coarsening_scheme', 2) sfdp_path = kwargs.get('sfdp_path', './bin/sfdp_osx') embeddings = np.ndarray(shape=(graph.number_of_nodes(), representation_size), dtype=np.float32) for subgraph, reversed_mapping in zip(subgraphs, reversed_mappings): count += 1 print ('Subgraph %d with %d nodes and %d edges' % (count, subgraph.number_of_nodes(), subgraph.number_of_edges())) if not subgraph.is_connected(): gc_single_model = baseline.skipgram_baseline(subgraph, scale=scale, num_paths=num_paths, path_length=path_length, iter_count=iter_count, representation_size=representation_size, window_size=window_size, report_loss=report_loss, progress_threshold=100000, alpha=alpha, min_alpha=min_alpha, sg=1, hs=hs) gc_model = [gc_single_model] else: if recursive_graphs is None: print ('Graph Coarsening...') recursive_graphs, recursive_merged_nodes = external_ec_coarsening(subgraph, sfdp_path) iter_counts = [iter_count for _ in range(len(recursive_graphs))] if hs == 1: gc_model = skipgram_coarsening_hs(recursive_graphs, recursive_merged_nodes, scale=scale, iter=iter_counts, num_paths=num_paths, path_length=path_length, representation_size=representation_size, window_size=window_size, report_loss=report_loss, progress_threshold=100000, lr_scheme=lr_scheme, alpha=alpha, min_alpha=min_alpha, sg=1, hs=1, sample=sample) else: print ('Training negative sampling model...') gc_model = skipgram_coarsening_neg(recursive_graphs, recursive_merged_nodes, scale=scale, iter=iter_counts, num_paths=num_paths, path_length=path_length, representation_size=representation_size, window_size=window_size, report_loss=report_loss, progress_threshold=100000, lr_scheme=lr_scheme, alpha=alpha, min_alpha=min_alpha, sample=sample, sg=1, hs=0) for ind, vec in enumerate(gc_model[-1].wv.syn0): real_ind = reversed_mapping[int(gc_model[-1].wv.index2word[ind])] embeddings[real_ind] = vec recursive_graphs = None return embeddings def gen_alpha(init_alpha, recursive_graphs, iter_counts): edge_counts = [graph.number_of_edges() for graph in recursive_graphs] total_iter_count = sum([edge_count * iter_count for edge_count, iter_count in zip(edge_counts, iter_counts)]) cur_iter_count, alpha_list = 0, [] for edge_count, iter_count in zip(edge_counts, iter_counts): cur_iter_count += edge_count * iter_count alpha_list.append(init_alpha * 1. * cur_iter_count / total_iter_count) return alpha_list def skipgram_coarsening_hs(recursive_graphs, recursive_merged_nodes, **kwargs): print (kwargs) print ('Start building Skip-gram + Hierarchical Softmax model on the coarsened graphs...') models = [] original_graph = recursive_graphs[0] levels = len(recursive_graphs) alpha = kwargs.get('alpha', 0.25) min_alpha = kwargs.get('min_alpha', 0.25) tmp_alpha_list = gen_alpha(alpha, recursive_graphs, kwargs['iter']) lr_scheme = kwargs.get('lr_scheme', "default") sample = kwargs.get('sample', 1e-3) # learning rate schemes: "default", "constant", "global_linear", "local_linear" if lr_scheme == 'default': alpha_list = [alpha for i in range(levels)] min_alpha_list = [min_alpha for i in range(levels)] if kwargs["lr_scheme"] == 'constant': alpha_list = [alpha for i in range(levels)] min_alpha_list = [alpha for i in range(levels)] elif kwargs["lr_scheme"] == 'local_linear': alpha_list = [alpha for alpha in tmp_alpha_list] min_alpha_list = [min_alpha for i in range(levels)] elif kwargs["lr_scheme"] == 'global_linear': alpha_list = [alpha for alpha in tmp_alpha_list] min_alpha_list = [min_alpha] min_alpha_list.extend([tmp_alpha_list[i] for i in range(levels - 1)]) scale = kwargs.get('scale', 1) if 'walks' in kwargs: walks = kwargs['walks'] for level in range(levels - 1, -1, -1): print ('Training on graph level %d...' % level) if scale == 1: edges, weights = recursive_graphs[level].get_edges() random.shuffle(edges) elif scale == -1: path_length = kwargs.get('path_length', 10) num_paths = kwargs.get('num_paths', 40) output = kwargs.get('output', 'default') edges = build_deepwalk_corpus(recursive_graphs[level], num_paths, path_length, output) # the coarest level if level == levels - 1: model = skipgram.Word2Vec_hs_loss(edges, sg=kwargs['sg'], size=kwargs['representation_size'], iter=kwargs['iter'][level], window=kwargs['window_size'], sample=sample, alpha=alpha_list[level], min_alpha=min_alpha_list[level]) else: model = skipgram.Word2Vec_hs_loss(None, sg=kwargs['sg'], size=kwargs['representation_size'], iter=kwargs['iter'][level], window=kwargs['window_size'], sample=sample, alpha=alpha_list[level], min_alpha=min_alpha_list[level]) # copy vocab / index2word from the coarser graph model.vocab = copy.deepcopy(models[-1].wv.vocab) model.index2word = copy.deepcopy(models[-1].wv.index2word) model.syn0 = copy.deepcopy(models[-1].wv.syn0) model.syn0.resize(recursive_graphs[level].number_of_nodes(), kwargs['representation_size']) model.syn0norm = None model.corpus_count = len(edges) cur_merged_nodes = [(node, merged_node) for node, merged_node in recursive_merged_nodes[level].iteritems() if node != merged_node] cur_merged_nodes = sorted(cur_merged_nodes, key=operator.itemgetter(1)) changed_merged_nodes = [] cur_merged_node, prev_node = -1, -1 node_pool = [] for node, merged_node in cur_merged_nodes: if merged_node == cur_merged_node: changed_merged_nodes.append((node, random.choice(node_pool))) node_pool.append(node) else: changed_merged_nodes.append((node, merged_node)) cur_merged_node = merged_node node_pool = [node, merged_node] prev_node = node cur_index = len(models[-1].mv.vocab) for node, merged_node in changed_merged_nodes: if node == merged_node: continue str_node, str_merged_node = str(node), str(merged_node) word_index = model.vocab[str_merged_node].index init_vec = model.syn0[word_index] model.add_word(str_node, str_merged_node, init_vec, cur_index) cur_index += 1 model.add_word(str_merged_node, str_merged_node, init_vec, cur_index) model.syn1 = np.zeros((len(model.vocab), model.layer1_size), dtype=np.float32) for i in range(len(models[-1].syn1)): model.syn1[i] = models[-1].syn1[i] model.syn0_lockf = np.ones(len(model.vocab), dtype=np.float32) model.train(edges) models.append(model) print ('Finish building Skip-gram model on the coarsened graphs.') return models def skipgram_coarsening_neg(recursive_graphs, recursive_merged_nodes, **kwargs): # print (kwargs) print ('Start building Skip-gram + Negative Sampling model on the coarsened graphs...') models = [] original_graph = recursive_graphs[0] levels = len(recursive_graphs) tmp_alpha_list = gen_alpha(kwargs.get('alpha', 0.025), recursive_graphs, kwargs['iter']) # learning rate schemes: "constant", "global_linear", "local_linear" if kwargs["lr_scheme"] == 'default': alpha_list = [kwargs['alpha'] for i in range(levels)] min_alpha_list = [kwargs['min_alpha'] for i in range(levels)] if kwargs["lr_scheme"] == 'constant': alpha_list = [kwargs['alpha'] for i in range(levels)] min_alpha_list = [kwargs['alpha'] for i in range(levels)] elif kwargs["lr_scheme"] == 'local_linear': alpha_list = [alpha for alpha in tmp_alpha_list] min_alpha_list = [kwargs['min_alpha'] for i in range(levels)] elif kwargs["lr_scheme"] == 'global_linear': alpha_list = [alpha for alpha in tmp_alpha_list] min_alpha_list = [kwargs['min_alpha']] min_alpha_list.extend([tmp_alpha_list[i] for i in range(levels - 1)]) scale = kwargs.get('scale', 1) sample = kwargs.get('sample', 1e-3) for level in range(levels - 1, -1, -1): print ('Training on graph level %d...' % level) # DeepWalk if scale == -1: path_length = kwargs.get('path_length', 10) num_paths = kwargs.get('num_paths', 40) output = kwargs.get('output', 'default') edges = build_deepwalk_corpus(recursive_graphs[level], num_paths, path_length, output) # use adjacency matrix elif scale == 1: edges, weights = recursive_graphs[level].get_edges() random.shuffle(edges) # the coarest level if level == levels - 1: model = Word2Vec(edges, size=kwargs['representation_size'], window=kwargs['window_size'], min_count=0, sample=sample, sg=1, hs=0, iter=kwargs['iter'][level], workers=20) else: model = Word2Vec(None, size=kwargs['representation_size'], window=kwargs['window_size'], min_count=0, sample=sample, sg=1, hs=0, iter=kwargs['iter'][level], workers=20) model.build_vocab(edges) model.reset_weights() # init model weights with the previous one prev_syn0 = {models[-1].wv.index2word[ind]: vec for ind, vec in enumerate(models[-1].wv.syn0)} prev_syn1neg = {models[-1].wv.index2word[ind]: vec for ind, vec in enumerate(models[-1].syn1neg)} word2index = {model.wv.index2word[ind]: ind for ind in range(recursive_graphs[level].number_of_nodes())} for ind in range(recursive_graphs[level].number_of_nodes()): word = model.wv.index2word[ind] if word in prev_syn0: model.wv.syn0[ind] = prev_syn0[word] model.syn1neg[ind] = prev_syn1neg[word] else: # if a is merged into b, then a should has identical weights in word2vec as b if int(word) in recursive_merged_nodes[level]: word_ind = word2index[word] merged_word = str(recursive_merged_nodes[level][int(word)]) model.wv.syn0[word_ind] = prev_syn0[merged_word] model.syn1neg[word_ind] = prev_syn1neg[merged_word] model.syn0_lockf = np.ones(len(model.wv.vocab), dtype=np.float32) model.train(edges) models.append(model) print ('Finish building Skip-gram model on the coarsened graphs.') return models class combine_files_iter: def __init__(self, file_list, length, path_length): self.file_list = file_list self.file_list_iter = iter(file_list) self.fp_iter = open(next(self.file_list_iter)) self.length = length self.path_length = path_length def __len__(self): return self.length def __iter__(self): for fname in self.file_list: for line in open(fname): yield line.split() # return self def next(self): try: result = next(self.fp_iter).split() except: try: self.fp_iter.close() self.fp_iter = open(next(self.file_list_iter)) result = next(self.fp_iter).split() except: raise StopIteration return result def build_deepwalk_corpus(G, num_paths, path_length, output, alpha=0): walks_filebase = output + '.walks' walk_files = serialized_walks.write_walks_to_disk(G, walks_filebase, num_paths=num_paths, path_length=path_length, alpha=alpha, rand=random.Random(random.randint(0, 2**31)), num_workers=20) return combine_files_iter(walk_files, G.number_of_nodes() * num_paths, path_length)
-
baseline.py
from gensim.models import Word2Vec import graph_coarsening import numpy as np def skipgram_baseline(graph, **kwargs): scale = kwargs.get('scale', -1) representation_size = kwargs.get('representation_size', 128) if scale == 1: edges, weights = graph.get_edges() else: path_length = kwargs.get('path_length', 40) num_paths = kwargs.get('num_paths', 80) output = kwargs.get('output', 'default') edges = graph_coarsening.build_deepwalk_corpus(graph, num_paths, path_length, output) if kwargs['hs'] == 0: print ('Training the Negative Sampling Model...') model = Word2Vec(edges, size=representation_size, window=kwargs['window_size'], min_count=0, sg=1, hs=0, iter=kwargs['iter_count'], negative=5, workers=20) else: print ('Training the Hierarchical Softmax Model...') model = Word2Vec(edges, size=kwargs['representation_size'], window=kwargs['window_size'], min_count=0, sg=1, hs=1, iter=kwargs['iter_count'], workers=20) print ('Finish training the Skip-gram model.') return model -
skipgram.py(train函数和其余gensim移入wv的模块都做了修改)
from __future__ import division # py3 "true division" import logging import sys import os import heapq import copy import numpy as np from timeit import default_timer from copy import deepcopy from collections import defaultdict import threading import itertools try: from queue import Queue, Empty except ImportError: from Queue import Queue, Empty from numpy import exp, log, dot, zeros, outer, random, dtype, float32 as REAL,\ uint32, seterr, array, uint8, vstack, fromstring, sqrt, newaxis,\ ndarray, empty, sum as np_sum, prod, ones, ascontiguousarray from gensim import utils, matutils # utility fnc for pickling, common scipy operations etc from gensim.models import Word2Vec from gensim.models.word2vec import Vocab from six import iteritems, itervalues, string_types from six.moves import xrange from types import GeneratorType import random logger = logging.getLogger(__name__) try: from gensim.models.word2vec_inner import train_batch_sg, train_batch_cbow from gensim.models.word2vec_inner import score_sentence_sg, score_sentence_cbow from gensim.models.word2vec_inner import FAST_VERSION, MAX_WORDS_IN_BATCH except ImportError: # failed... fall back to plain numpy (20-80x slower training than the above) FAST_VERSION = -1 MAX_WORDS_IN_BATCH = 10000 # modified hierarchical softmax model based on Gensim's implementation class Word2Vec_hs_loss(Word2Vec): def __init__(self, sentences=None, **kwargs): self.inner_node_index_map = {} kwargs["hs"] = 1 kwargs["alpha"] = kwargs.get("alpha", 0.025) kwargs["min_alpha"] = kwargs.get("min_alpha", 0.001) kwargs["min_count"] = 0 kwargs["negative"] = 0 kwargs["sample"] = kwargs.get("sample", 1e-3) kwargs["workers"] = kwargs.get("workers", 20) super(self.__class__, self).__init__(sentences, **kwargs) # add a word as the child of current word in the coarser graph def add_word(self, word, parent_word, emb, cur_index): fake_vocab_size = int(1e7) word_index = len(self.wv.vocab) inner_node_index = word_index - 1 parent_index = self.wv.vocab[parent_word].index # add in the left subtree if word != parent_word: self.wv.vocab[word] = Vocab(index=word_index, count=fake_vocab_size-word_index,sample_int=(2**32)) if emb is not None: self.wv.syn0[cur_index] = emb else: self.wv.syn0[cur_index] = self.wv.syn0[parent_index] # the node in the coarsened graph serves as an inner node now self.wv.index2word.append(word) self.wv.vocab[word].code = array(list(self.wv.vocab[parent_word].code) + [0], dtype=uint8) self.wv.vocab[word].point = array(list(self.wv.vocab[parent_word].point) + [inner_node_index], dtype=uint32) self.inner_node_index_map[parent_word] = inner_node_index else: if emb is not None: self.wv.syn0[parent_index] = emb self.wv.vocab[word].code = array(list(self.wv.vocab[word].code) + [1], dtype=uint8) self.wv.vocab[word].point = array(list(self.wv.vocab[word].point) + [self.inner_node_index_map[word]], dtype=uint32) def train(self, sentences, total_words=None, word_count=0, total_examples=None, queue_factor=2, report_delay=0.1, **kwargs): """ Update the model's neural weights from a sequence of sentences (can be a once-only generator stream). For Word2Vec, each sentence must be a list of unicode strings. (Subclasses may accept other examples.) To support linear learning-rate decay from (initial) alpha to min_alpha, either total_examples (count of sentences) or total_words (count of raw words in sentences) should be provided, unless the sentences are the same as those that were used to initially build the vocabulary. """ self.loss = {} if FAST_VERSION < 0: import warnings warnings.warn("C extension not loaded for Word2Vec, training will be slow. " "Install a C compiler and reinstall gensim for fast training.") self.neg_labels = [] if self.negative > 0: # precompute negative labels optimization for pure-python training self.neg_labels = zeros(self.negative + 1) self.neg_labels[0] = 1. logger.info( "training model with %i workers on %i vocabulary and %i features, " "using sg=%s hs=%s sample=%s negative=%s", self.workers, len(self.wv.vocab), self.layer1_size, self.sg, self.hs, self.sample, self.negative) if not self.wv.vocab: raise RuntimeError("you must first build vocabulary before training the model") if not hasattr(self.wv, 'syn0'): raise RuntimeError("you must first finalize vocabulary before training the model") if total_words is None and total_examples is None: if self.corpus_count: total_examples = self.corpus_count logger.info("expecting %i sentences, matching count from corpus used for vocabulary survey", total_examples) else: raise ValueError("you must provide either total_words or total_examples, to enable alpha and progress calculations") job_tally = 0 if self.iter > 1: sentences = utils.RepeatCorpusNTimes(sentences, self.iter) total_words = total_words and total_words * self.iter total_examples = total_examples and total_examples * self.iter def worker_loop(): """Train the model, lifting lists of sentences from the job_queue.""" work = matutils.zeros_aligned(self.layer1_size, dtype=REAL) # per-thread private work memory neu1 = matutils.zeros_aligned(self.layer1_size, dtype=REAL) jobs_processed = 0 while True: job = job_queue.get() if job is None: progress_queue.put(None) break # no more jobs => quit this worker sentences, alpha = job tally, raw_tally = self._do_train_job(sentences, alpha, (work, neu1)) progress_queue.put((len(sentences), tally, raw_tally)) # report back progress jobs_processed += 1 # logger.debug("worker exiting, processed %i jobs", jobs_processed) def job_producer(): """Fill jobs queue using the input `sentences` iterator.""" job_batch, batch_size = [], 0 pushed_words, pushed_examples = 0, 0 next_alpha = self.alpha job_no = 0 for sent_idx, sentence in enumerate(sentences): sentence_length = self._raw_word_count([sentence]) # can we fit this sentence into the existing job batch? if batch_size + sentence_length <= self.batch_words: # yes => add it to the current job job_batch.append(sentence) batch_size += sentence_length else: # no => submit the existing job #logger.debug( # "queueing job #%i (%i words, %i sentences) at alpha %.05f", # job_no, batch_size, len(job_batch), next_alpha) job_no += 1 job_queue.put((job_batch, next_alpha)) # update the learning rate for the next job if self.min_alpha < next_alpha: if total_examples: # examples-based decay pushed_examples += len(job_batch) progress = 1.0 * pushed_examples / total_examples else: # words-based decay pushed_words += self._raw_word_count(job_batch) progress = 1.0 * pushed_words / total_words next_alpha = self.alpha - (self.alpha - self.min_alpha) * progress next_alpha = max(self.min_alpha, next_alpha) # add the sentence that didn't fit as the first item of a new job job_batch, batch_size = [sentence], sentence_length # add the last job too (may be significantly smaller than batch_words) if job_batch: # logger.debug( # "queueing job #%i (%i words, %i sentences) at alpha %.05f", # job_no, batch_size, len(job_batch), next_alpha) job_no += 1 job_queue.put((job_batch, next_alpha)) if job_no == 0 and self.train_count == 0: logger.warning( "train() called with an empty iterator (if not intended, " "be sure to provide a corpus that offers restartable " "iteration = an iterable)." ) # give the workers heads up that they can finish -- no more work! for _ in xrange(self.workers): job_queue.put(None) logger.debug("job loop exiting, total %i jobs", job_no) # buffer ahead only a limited number of jobs.. this is the reason we can't simply use ThreadPool :( job_queue = Queue(maxsize=queue_factor * self.workers) progress_queue = Queue(maxsize=(queue_factor + 1) * self.workers) workers = [threading.Thread(target=worker_loop) for _ in xrange(self.workers)] unfinished_worker_count = len(workers) workers.append(threading.Thread(target=job_producer)) for thread in workers: thread.daemon = True # make interrupting the process with ctrl+c easier thread.start() example_count, trained_word_count, raw_word_count = 0, 0, word_count start, next_report = default_timer() - 0.00001, 1.0 prev_example_count = 0 while unfinished_worker_count > 0: report = progress_queue.get() # blocks if workers too slow if report is None: # a thread reporting that it finished unfinished_worker_count -= 1 # logger.info("worker thread finished; awaiting finish of %i more threads", unfinished_worker_count) continue examples, trained_words, raw_words = report job_tally += 1 # update progress stats example_count += examples trained_word_count += trained_words # only words in vocab & sampled raw_word_count += raw_words # log progress once every report_delay seconds elapsed = default_timer() - start if elapsed >= next_report: next_report = elapsed + report_delay # all done; report the final stats elapsed = default_timer() - start logger.info( "training on %i raw words (%i effective words) took %.1fs, %.0f effective words/s", raw_word_count, trained_word_count, elapsed, trained_word_count / elapsed) if job_tally < 10 * self.workers: logger.warn("under 10 jobs per worker: consider setting a smaller `batch_words' for smoother alpha decay") # check that the input corpus hasn't changed during iteration if total_examples and total_examples != example_count: logger.warn("supplied example count (%i) did not equal expected count (%i)", example_count, total_examples) if total_words and total_words != raw_word_count: logger.warn("supplied raw word count (%i) did not equal expected count (%i)", raw_word_count, total_words) self.train_count += 1 # number of times train() has been called self.total_train_time += elapsed self.clear_sims() return trained_word_count