使用SVM/k-NN模型实现手写数字多分类 - 清华大学《机器学习实践与应用》22春-周作业
0 Contents
-
-
- 1.1 多分类SVM主要思想
-
- 1.1.1 一对一SVM分类(OvO-SVM)
- 1.1.2 一对多SVM分类(OvR-SVM)
- 1.2 实验设计及伪代码
-
- 1.2.1 实验目的概述
- 1.2.2 实验模型整体设计
- 1.2.3 多分类伪代码及解释
- 1.2.4 完整代码实现
- 1.3 测试结果及分析
- 1.4 其他实验体会
-
1.1 多分类SVM主要思想
SVM模型处理分类问题建立在使用一个超平面分割两类数据,根据几何位态分别赋予标签 + 1 +1 +1和 − 1 -1 −1以区分不同样本。目前较为通用的基于SVM的多分类方法一般采用集成学习思想,包括:一对一分类(One vs. One, OvO)和一对多分类(One vs. Rest, OvR)。
1.1.1 一对一SVM分类(OvO-SVM)
假设输入数据 { ( x i , y i ) } i = 1 N \{(x_i,y_i)\}_{i=1}^{N} {(xi,yi)}i=1N中 y i ∈ { 1 , ⋅ , M } y_i \in \{1,\cdot,M\} yi∈{1,⋅,M}个不同分类。OvO-SVM对任何无序类别组合( a a a, b b b)都训练且仅训练一个二分类SVM(共 M ( M − 1 ) / 2 M(M-1)/2 M(M−1)/2个子分类器),并求解如下优化问题:
min w ( a b ) , b ( a b ) , ξ ( a b ) 1 2 w ( a b ) T w ( a b ) + C ∑ i N ξ i ( a b ) , s . t . I a { y i } ( w ( a b ) T x i + b ( a b ) ) ≥ 1 − ξ i ( a b ) , ξ i ( a b ) ≥ 0 , ∀ i = 1 , … , N \min_{w^{(ab)},b^{(ab)},\xi^{(ab)}} \frac{1}{2}{w^{(ab)}}^Tw^{(ab)} + C \sum_i^N \xi^{(ab)}_i,\ s.t. \mathbb I_{a}\{y_i\}({w^{(ab)}}^Tx_i+b^{(ab)}) \geq 1 - \xi^{(ab)}_i,\ \xi^{(ab)}_i \geq 0,\ ∀ i=1,\dots,N w(ab),b(ab),ξ(ab)min21w(ab)Tw(ab)+Ci∑Nξi(ab), s.t.Ia{yi}(w(ab)Txi+b(ab))≥1−ξi(ab), ξi(ab)≥0, ∀i=1,…,N
其中, I a { y i } \mathbb I_a\{y_i\} Ia{yi}是一个双极性指示函数,当 y i = a y_i=a yi=a时(正类),值为 + 1 +1 +1,反之当 y i ≠ a y_i \neq a yi=a时(负类, y i = b y_i=b yi=b),值为 − 1 -1 −1.
最后,对于样本标签的预测,可以通过在所有分类器的预测结果中分别计数预测成各类标签的个数,并基于投票策略得出集成结果。
当然,也可以考虑先统计每个标签下样本出现的古典概率 P ( Y = m t ∣ X = x i ) P(Y=m_t|X=x_i) P(Y=mt∣X=xi),并基于贝叶斯决策求解样本的后验分布:
y ^ i = arg max t P ( Y = m t ∣ X = x i ) = arg max t P ( Y = m t ) P ( X = x i ∣ Y = m t ) ∑ q P ( Y = m q ) P ( X = x i ∣ Y = m q ) , m t = 1 , … , M \hat y_i = \arg\max_t P(Y=m_t|X=x_i) = \arg\max_t \frac{P(Y=m_t)P(X=x_i | Y=m_t)}{\sum_qP(Y=m_q)P(X=x_i | Y=m_q)},\ m_t=1,\dots,M y^i=argtmaxP(Y=mt∣X=xi)=argtmax∑qP(Y=mq)P(X=xi∣Y=mq)P(Y=mt)P(X=xi∣Y=mt), mt=1,…,M
本题代码设计中采用投票策略实现。
1.1.2 一对多SVM分类(OvR-SVM)
基本符号定义同1.1.1节。和OvO-SVM相比,OvR-SVM对任何类别 a a a训练一个二分类SVM,并求解:
min w ( a ) , b ( a ) , ξ ( a ) 1 2 w ( a ) T w ( a ) + C ∑ i N ξ i ( a ) , s . t . I a { y i } ( w ( a ) T x i + b ( a ) ) ≥ 1 − ξ i ( a ) , ξ i a ≥ 0 , ∀ i = 1 , … , N \min_{w^{(a)},b^{(a)},\xi^{(a)}} \frac{1}{2}{w^{(a)}}^Tw^{(a)} + C \sum_i^N \xi^{(a)}_i,\ s.t. \mathbb I_{a}\{y_i\}({w^{(a)}}^Tx_i+b^{(a)}) \geq 1 - \xi^{(a)}_i,\ \xi^{a}_i \geq 0,\ ∀ i=1,\dots,N w(a),b(a),ξ(a)min21w(a)Tw(a)+Ci∑Nξi(a), s.t.Ia{yi}(w(a)Txi+b(a))≥1−ξi(a), ξia≥0, ∀i=1,…,N
此时,通过在每个类别标签对应的分类器上分别对样本进行预测,可以得到 N N N个分类结果。
- 理想情况下,应当只有一个分类器预测为正类,其余为负类。此时样本标签即为正类对应的标签。
- 如果有多个标签被预测为正例,可以从中选择置信最高的那个类别,其置信可以从其数值强度/到超平面距离来判断,如预测类别a为 + 1.37 +1.37 +1.37,而预测类别 b b b为 + 3.5 +3.5 +3.5。那么显然预测为b是更加合理的选择。
- 如果没有类别被预测为正例,说明样本没有被判断为任何类别,此时依然可以考虑置信思路,对每个类别计算置信的负分,并选择惩罚最小的那个类别作为当前预测的正例标签。
本题代码设计中仅处理理想情况的逻辑,并对异常预测结果随机选择预测的类别。
1.2 实验设计及伪代码
1.2.1 实验目的概述
- 实现多分类SVM的主要逻辑,包括OvR-SVM和OvO-SVM.
- 比较k-NN和SVM模型在分类任务上的7项指标。
- 模型参数选择:
- k-NN:测试k=1,3,10时对应的模型表现
- SVM:测试线性核、RBF核(0.1,5,10,50,100)
- 测试指标
- 训练时间(s)
- 训练过程占用内存(MB)
- 训练集上分类正确率(%)
- 测试时间(s)
- 测试过程占用内存(MB)
- 测试集上分类正确率(%)
- 模型参数规模(MB)
- 模型参数选择:
1.2.2 实验模型整体设计
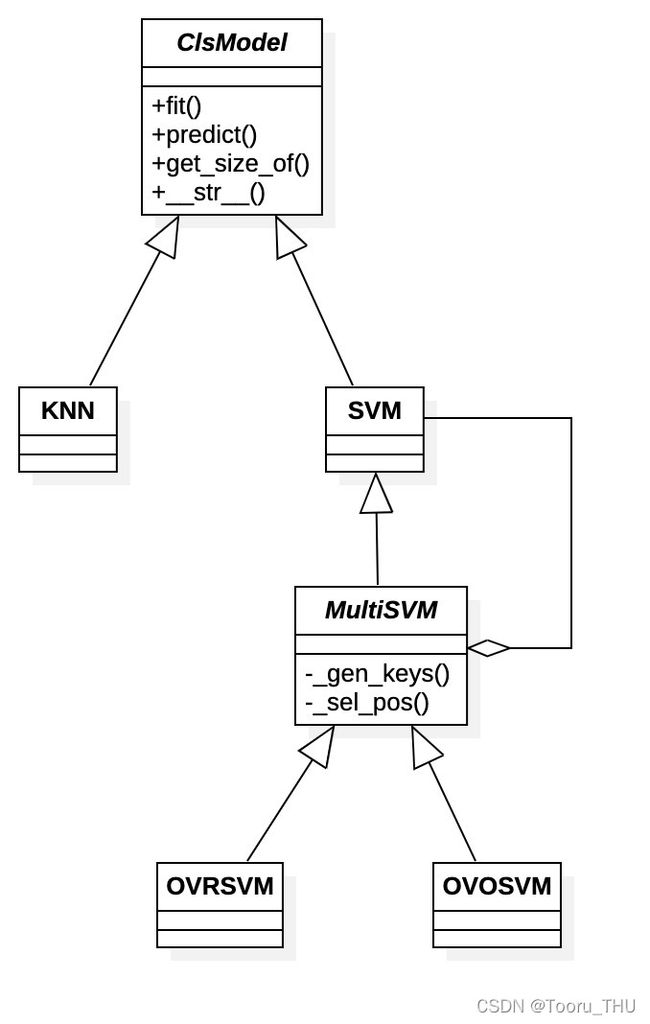
我们使用软件设计模式中的UML类图描述实验中模型的封装与组织关系,如下。
其中,
- ClsModel定义为分类模型的抽象超类,描述分类模型具备的基本逻辑接口,并支持对参数规模统计。
- KNN:向下封装课程脚本KNN.py相关逻辑,向上提供统一的分类接口实现。
- SVM:向下封装课程脚本svmMLiA.py相关逻辑,向上提供统一的分类接口实现。
- MultiSVM:基于SVM构造的多分类SVM抽象类,内部动态构造多个子SVM实体,描述子分类器构造规则(gen_keys)和类别判断规则(sel_pos)的抽象接口。
- OVRSVM:实现一对多SVM分类的实体类
- OVOSVM:实现一对一SVM分类的实体类
1.2.3 多分类伪代码及解释
根据上节对功能模块的职责划分,我们进一步解释对MultiSVM、OVRSVM、OVOSVM的伪代码实现逻辑。
- MultiSVM
def fit(data,labels):
svm_dict = {}
for key,complete_data in gen_key(data,labels):
shuffle complete_data.
construct svm by complete data.
svm_dict[k] = svm
def predict(data):
votes = []
for each label in label_set:
for key,inferred_label in sel_pos(label):
svm_dict[k].predict(data)
prediction is positive or negative? choose the corresponding inferred label.
append the chosen label to votes
finally predict by votes
其中,fit方法根据gen_key确定的分类器的输入参数(包括某一策略下标志键和对应的完整数据),打乱数据,并执行SVM的训练。predict方法对每种标签可能性遍历,并根据sel_pos方法选择的每类标签能够充分预测所需的标志键key和基于预测结果最终应当被推断的标签二元组inferred_label(即positive class和negative class对应的标签构成的二元组),选择对应的子SVM执行预测,并将结果投入到votes中去。最后基于全体投票得出准确的预测标签。
基于上述抽象的多分类框架设计,考虑OvR和OVO具体策略的差异,分别实现对应的gen_key和sel_pos。
- OVRSVM
def gen_keys(data,labels):
for each label in label_set:
assign label to +1, other labels to -1
yield label, transformed data
def sel_pos(label):
return [(label,(label,None))]
根据OVRSVM的思路,gen_key需要给出每个标签作为正例,其余标签作为负例的 M M M中情形,并对完整数据进行双极性标注返回。
sel_pos则只赋予正类对应真实的标签,负类不执行标签推断。
- OVOSVM
def _gen_keys(data,labels):
for (label_a,label_b) in label_set:
assign label_a to +1, label_b to -1
yield 'label_a - label_b', transformed data
def sel_pos(label):
return ['label - label_x',(label,label_x) for x in other_labels]
OVOSVM在gen_key中对每一个无序的标签二元组(label_a,label_b)进行标注,其中label_a及其对应数据标注为正类,label_b为负类
sel_pos则分别对应正负类都推断真实的标签a和b。
1.2.4 完整代码实现
本实验的完整模型实现如下。
'''
implemented classes of classifiers.
'''
class ClsModel:
def __init__(self,model_name='anonymous',num_cls=10):
self.model_name = model_name
self.num_cls = num_cls
self.param_lst = []
def fit(self,data,labels):
'''
fit cls model with train data.
:param data: ndarray[n × m], n - samples, m - dimension
:param labels: ndarray[1 × m], m - dimension
:return:
'''
raise NotImplementedError
def predict(self,data):
'''
predict test samples of their labels.
:param data: ndarray[n × m], n - samples, m - dimension
:return: ndarray[1 × m] - labels
'''
raise NotImplementedError
def get_size_of(self):
return sum([sys.getsizeof(ele) for ele in self.param_lst])
def __str__(self):
return 'ClsModel-' + self.model_name if self.model_name == 'anonymous' else 'ClsModel'
class KNN(ClsModel):
def __init__(self,k=3,**kwargs):
super(KNN, self).__init__(**kwargs)
self.k = k
def fit(self,data,labels):
self.data = copy.deepcopy(data) # copy since we require data NOT be destroyed, mem anal as well.
self.labels = copy.deepcopy(labels)
self.param_lst.extend([self.data,self.labels])
def predict(self,data):
return np.array([m_knn.classify0(inX=row_data,dataSet=self.data,labels=self.labels,k=self.k) for row_data in data])
def __str__(self):
return 'KNN-' + self.model_name
class SVM(ClsModel):
def __init__(self,C=200,toler=1e-4,maxIter=40,kTup = ('lin',0),**kwargs):
super(SVM, self).__init__(**kwargs)
self.C = C
self.toler =toler
self.maxIter = maxIter
self.kTup = kTup
self.param_lst.extend([self.C,self.toler,self.maxIter,self.kTup])
def fit(self,data,labels):
self.b, alphas = m_svm.smoPK(data, labels, self.C, self.toler, self.maxIter, self.kTup)
datMat = np.mat(data)
labelMat = np.mat(labels).transpose()
svInd = np.nonzero(alphas.A > 0)[0]
self.sVs = datMat[svInd]
self.labelSV = labelMat[svInd]
self.alphas = alphas[svInd]
self.param_lst.extend([self.b,self.alphas,self.sVs,self.labelSV])
def predict(self,data):
return np.array([m_svm.kernelTrans(self.sVs, row_data, self.kTup).T * np.multiply(self.labelSV,self.alphas) + self.b for row_data in np.mat(data)])
def __str__(self):
return 'SVM-' + self.model_name if self.model_name == 'anonymous' else 'SVM'
class MultiSVM(SVM):
def __init__(self,**kwargs):
super(MultiSVM, self).__init__(**kwargs)
self.param_dict = kwargs
self.svm_dict = {}
def fit(self,data,labels):
# init sub-SVM model.
for k,cdata in self._gen_keys(data,labels):
rnd_idx = list(range(cdata.shape[0]))
random.shuffle(rnd_idx)
svm = SVM(**self.param_dict)
svm.fit(cdata[rnd_idx][:,:-1],cdata[rnd_idx][:,-1].reshape(-1))
self.svm_dict[k] = svm
def predict(self,data):
preds = []
for row_data in data:
lb2cnt = {}
max_lb = None
max_cnt = 0
for label in range(self.num_cls):
for k,inf_lb in zip(*self._sel_pos(label)):
pred = self.svm_dict[k].predict(row_data.reshape(1, -1))
pred = 0 if pred > 0 else 1 # lb2idx.
if inf_lb[pred] is not None:
lb2cnt[inf_lb[pred]] = lb2cnt.get(inf_lb[pred],0) + 1
if lb2cnt[inf_lb[pred]] > max_cnt:
max_cnt = lb2cnt[inf_lb[pred]]
max_lb = inf_lb[pred]
# assert max_lb is not None
preds.append(max_lb if max_lb is not None else random.choice(range(self.num_cls)))
return preds
def _gen_keys(self,data,labels):
raise NotImplementedError
def _sel_pos(self,label):
raise NotImplementedError
def get_size_of(self):
return sum([v.get_size_of() for k,v in self.svm_dict.items()])
def __str__(self):
return 'MultiSVM-' + self.model_name if self.model_name == 'anonymous' else 'MultiSVM'
class OVRSVM(MultiSVM):
def _gen_keys(self,data,labels):
cdata = np.concatenate([data, labels.reshape(-1, 1)], axis=1)
for pos_cls in range(self.num_cls):
pos_data = cdata[cdata[:, -1] == pos_cls]
neg_data = cdata[cdata[:, -1] != pos_cls]
pos_data[:, -1] = 1
neg_data[:, -1] = -1
yield pos_cls, np.concatenate([pos_data, neg_data], axis=0)
def _sel_pos(self,label):
return [label],[(label,None)]
def __str__(self):
return 'OVRSVM-' + self.model_name if self.model_name == 'anonymous' else 'OVRSVM'
class OVOSVM(MultiSVM):
def _gen_keys(self, data, labels):
cdata = np.concatenate([data, labels.reshape(-1, 1)], axis=1)
for pos_cls in range(self.num_cls):
for neg_cls in range(pos_cls+1,self.num_cls):
pos_data = cdata[cdata[:, -1] == pos_cls]
neg_data = cdata[cdata[:, -1] == neg_cls]
pos_data[:, -1] = 1
neg_data[:, -1] = -1
yield '{}-{}'.format(pos_cls,neg_cls), np.concatenate([pos_data, neg_data], axis=0)
def _sel_pos(self, label):
return ['{}-{}'.format(label,num) for num in range(label+1,self.num_cls)], [(label,num) for num in range(label+1,self.num_cls)]
def __str__(self):
return 'OVOSVM-' + self.model_name if self.model_name == 'anonymous' else 'OVOSVM'
1.3 测试结果及分析
实验中,我们对1.2.1节中提及的7个实验指标和11种实验模型进行完全测试,测试代码逻辑如下。
'''
eval routine.
'''
def eval_one_stop(X_train,y_train,X_test,y_test,cls_model):
train_time = 0.
test_time = 0.
train_mem = 0.
test_mem = 0.
train_acc = 0.
test_acc = 0.
# train.
train_mem = sum(memory_profiler.memory_usage())
train_time = time.time()
cls_model.fit(X_train,y_train)
train_time = time.time() - train_time
train_mem = sum(memory_profiler.memory_usage()) - train_mem
train_acc = np.sum(cls_model.predict(X_train) == y_train) / X_train.shape[0]
# test.
test_mem = sum(memory_profiler.memory_usage())
test_time = time.time()
test_acc = np.sum(cls_model.predict(X_test) == y_test) / X_test.shape[0]
test_time = time.time() - test_time
test_mem = sum(memory_profiler.memory_usage()) - test_mem
return {
'train_time':train_time,
'train_mem':train_mem if train_mem > 0 else 0.,
'train_acc':train_acc,
'test_time':test_time,
'test_mem':test_mem if test_mem > 0 else 0.,
'test_acc':test_acc,
'storage':cls_model.get_size_of() / 1024 / 1024,
}
'''
routine.
'''
def routine1():
cls_knn_k1 = KNN(k=1)
cls_knn_k3 = KNN(k=3)
cls_knn_k10 = KNN(k=10)
cls_knn_k20 = KNN(k=20)
cls_svm_ovr_lin = OVRSVM(num_cls=10,kTup=('lin',0))
cls_svm_ovr_rbf01 = OVRSVM(num_cls=10, kTup=('rbf', 0.1))
cls_svm_ovr_rbf5 = OVRSVM(num_cls=10, kTup=('rbf', 5))
cls_svm_ovr_rbf10 = OVRSVM(num_cls=10, kTup=('rbf', 10))
cls_svm_ovr_rbf50 = OVRSVM(num_cls=10, kTup=('rbf', 50))
cls_svm_ovr_rbf100 = OVRSVM(num_cls=10, kTup=('rbf', 100))
cls_svm_ovo_lin = OVOSVM(num_cls=10,kTup=('lin',0))
cls_svm_ovo_rbf01 = OVOSVM(num_cls=10, kTup=('rbf', 0.1))
cls_svm_ovo_rbf5 = OVOSVM(num_cls=10, kTup=('rbf', 5))
cls_svm_ovo_rbf10 = OVOSVM(num_cls=10, kTup=('rbf', 10))
cls_svm_ovo_rbf50 = OVOSVM(num_cls=10, kTup=('rbf', 50))
cls_svm_ovo_rbf100 = OVOSVM(num_cls=10, kTup=('rbf', 100))
cls_models = [cls_knn_k1,cls_knn_k3,cls_knn_k10,cls_knn_k20,cls_svm_ovr_lin,cls_svm_ovo_lin,cls_svm_ovr_rbf01,cls_svm_ovo_rbf01,cls_svm_ovr_rbf5,cls_svm_ovo_rbf5,cls_svm_ovr_rbf10,cls_svm_ovo_rbf10,cls_svm_ovr_rbf50,cls_svm_ovo_rbf50,cls_svm_ovr_rbf100,cls_svm_ovo_rbf100]
cls_model_names = ['KNN@1','KNN@3','KNN@10','KNN@20','OVR-SVM-lin','OVO-SVM-lin','OVR-SVM-rbf0.1','OVO-SVM-rbf0.1','OVR-SVM-rbf5','OVO-SVM-rbf5','OVR-SVM-rbf10','OVO-SVM-rbf10','OVR-SVM-rbf50','OVO-SVM-rbf50','OVR-SVM-rbf100','OVO-SVM-rbf100']
metrics = ['train_time','train_mem','train_acc','test_time','test_mem','test_acc','storage']
X_train, y_train, X_test, y_test = load_and_split()
pddata = []
for idx,(cls_model,cls_model_name) in enumerate(zip(cls_models,cls_model_names)):
res_dict = eval_one_stop(X_train,y_train,X_test,y_test,cls_model)
pddata.append([res_dict[k] for k in metrics]) # ordered stream.
print('eval {} completed.'.format(cls_model))
# display&save after one completion.
anal_table = pd.DataFrame(data=np.array(pddata),columns=metrics,index=cls_model_names[:idx+1])
print(anal_table)
anal_table.to_csv('./fig/anal-all.csv')
对应实验结果如下。
从上述实验数据中可以得出多方面结论,总结如下。
- 训练时间上看,KNN方法具有极低的时间复杂度,这是因为其训练过程只是简单存储训练数据而没有显著的计算过程,相反,SVM方法则需要 O ( N 2 ) O(N^2) O(N2)的复杂度对输入数据遍历,并优化寻找其支持向量,RBF核的SVM由于还涉及数据矩阵的空间映射,耗时更长。此外,OvR方法训练耗时都普遍长于OvO方法,因为前者训练过程每个分类器都需要全部数据进行预测,需要执行 N 2 N^2 N2次计算,而OvO则仅执行成对的正负例训练,且总执行次数为 N ( N − 1 ) / 2 N(N-1)/2 N(N−1)/2次。
- 训练内存和测试内存上看,由于python内存申请的特性,导致各类方法的内存测定并不准确,但可以注意到KNN在训练和测试过程中几乎没有明显创建新的缓冲区,可以直接在数据空间上比较确定预测标签,效果较好,而采用OvO或OvR的SVM往往需要借助新的空间申请,并且新空间规模随模型参数变化。同时也可以看出,OvR虽然仅训练N个分类器却调用了更多的辅助空间,说明其空间占用更多来自于因完整数据规模较大造成特征向量规模成比例的增长。
- 训练和测试正确率上看,首先KNN方法表现总体优于SVM方法,因为相同的数字在书写像素格上往往具有较大面积的公有区域,使得简单的逐像素对比具有明显优势。同时,从参数上看,随着参数k的增加,训练集表现有所下降,但测试集表现先上升后下降,说明了模型从欠拟合到过拟合的变化过程。此外,对于SVM的线性核与RBF核对比上可以看出,RBF的精度表现普遍更高,说明数据在一定程度上是线性不可分的,手写数字的空间分布特征造成以径向基方式定义相关性会有利于不同数字样本的聚类特性。最后,对比OvO和OvR可以发现,后者的正确率表现往往不及前者,因为其存在类别不平衡问题,负例样本过多使得真实标签往往很难被正确预测出来。
- 测试时间上看,KNN具有相对稳定且较长的测试耗时,这与其较快的训练时间形成鲜明对比,而线性SVM由于前期已经对支持向量进行了筛选,该过程时间有明显下降,但另一方面,采用RBF核具有更坏的时间表现,说明主要的耗时更多发生在数据空间的映射上,这在需要训练 O ( N ~ N 2 ) O(N~N^2) O(N~N2)个分类器的多分类场景下表现更加突出。
- 模型参数规模上,KNN需要保存所有原始训练数据,占用参数最多,多分类SVM虽然需要保存所有子分类器的所有支持向量相关参数,但总体上参数规模依然相比KNN下降了2-3个量级。此外,OvR由于仅训练了N个分类器,参数规模相比OvO下降了近78%。
1.4 其他实验体会
- 良好的封装和代码规范。本实验中涉及较多的模型定义和逻辑调用,通过良好封装和抽象的类层次可以有效提高代码的可读性,减小冗余逻辑,便于错误调试和面向切面(AOP)的逻辑扩充。
- 基于toy data的预部署。本次实验耗时相对较长(上述完整数据在个人PC机跑完需要10h+),首先对在小规模数据上调试全部代码流程无误对实验效率很有帮助。例如实验中可以先设定num_cls=3仅对(0,1,2)三个数字的10个样本进行完整evaluation逻辑的执行,并通过断点调试和程序插桩对代码中可能存在的内存异常和逻辑错误及时纠正。