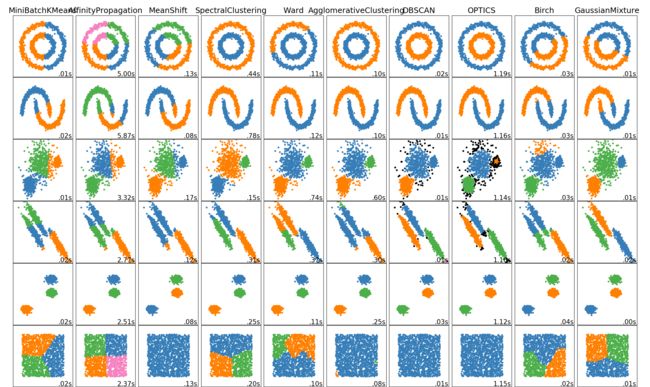
Scikit-learn:聚类clustering
http://blog.csdn.net/pipisorry/article/details/53185758
不同聚类效果比较
sklearn不同聚类示例比较
不同聚类综述
| Method name | Parameters | Scalability | Usecase | Geometry (metric used) |
|---|---|---|---|---|
| K-Means | number of clusters | Very large n_samples, medium n_clusters withMiniBatch code |
General-purpose, even cluster size, flat geometry, not too many clusters | Distances between points |
| Affinity propagation | damping, sample preference | Not scalable with n_samples | Many clusters, uneven cluster size, non-flat geometry | Graph distance (e.g. nearest-neighbor graph) |
| Mean-shift | bandwidth | Not scalable with n_samples |
Many clusters, uneven cluster size, non-flat geometry | Distances between points |
| Spectral clustering | number of clusters | Medium n_samples, small n_clusters |
Few clusters, even cluster size, non-flat geometry | Graph distance (e.g. nearest-neighbor graph) |
| Ward hierarchical clustering | number of clusters | Large n_samples and n_clusters |
Many clusters, possibly connectivity constraints | Distances between points |
| Agglomerative clustering | number of clusters, linkage type, distance | Large n_samples and n_clusters |
Many clusters, possibly connectivity constraints, non Euclideandistances | Any pairwise distance |
| DBSCAN | neighborhood size | Very large n_samples, medium n_clusters |
Non-flat geometry, uneven cluster sizes | Distances between nearest points |
| Gaussian mixtures | many | Not scalable | Flat geometry, good for density estimation | Mahalanobis distances to centers |
| Birch | branching factor, threshold, optional global clusterer. | Large n_clusters and n_samples |
Large dataset, outlier removal, data reduction. | Euclidean distance between points |
DBSCAN聚类
代码示例
def Dist(x, y): from geopy import distance return distance.vincenty(x, y).meters
import pickle, subprocess, pwd import pandas as pd import numpy as np import matplotlib.pyplot as plt from sklearn.cluster import DBSCAN df = pd.read_pickle(os.path.join(CWD, 'middlewares/df.pkl')) ll = df[['longitude', 'latitude']].values x, y = ll[:, 0], ll[:, 1] print('starting dbsan...')
dbscaner = DBSCAN(eps=DBSCAN_R, min_samples=DBSCAN_MIN_S, metric=Dist, n_jobs=-1).fit(ll) pickle.dump(dbscaner, open(os.path.join(CWD, 'middlewares/dbscaner.pkl'), 'wb')) print('dbsan dumping end...')dbscaner = pickle.load( open(os.path.join(CWD, 'middlewares/dbscaner.pkl'), 'rb'))labels = dbscaner.labels_ # print(set(labels))colors = plt.cm.Spectral(np.linspace( 0, 1, len( set(labels)))) for k, col in zip( set(labels), colors) : marker = '.' if k == - 1 : col = 'k' marker = 'x' inds_k = labels == k plt.scatter(x[inds_k], y[inds_k], marker =marker, color =col) if pwd.getpwuid(os.geteuid()).pw_name == 'piting' : plt.savefig( './1.png') elif pwd.getpwuid(os.geteuid()).pw_name == 'pipi' : plt.show()
[DBSCAN]
皮皮blog
from: sklearn:聚类clustering
ref: