Lecture 2——Basics of data processing
本文图片来自于学习视频——新一代测序技术数据分析第二讲
Lecture 2——Basics of data processing
Review Lecutre 1
Outline
Date analysis workflow
Sequence qualify evaluation
Phred scores
NGS error rates
Alignment
Smith-Waterman algorithm
Theories on short reads alignment
Suffix free, indexing, and Burrows-Wheeler transformation
Comparison of different aligners
Data formats
FASTQ, SAM, pileups, VCF
Data visualization
Genome Browsers, IGV,…
Date analysis workflow
HiSeq 2000 200G run
Image data: 32TB
Intensity Data: 2TB
Base call/quality score data: 250GB
Alignment output: 6TB(3TB), 1.2TB after intermediate files removed

Major steps for secondary analysis
Raw data —— QC Filter —— Alignment —— Annotation
Sequence quality
Base quality
For every nucleotide
Reported by the sequencer
Mapping quality (alignment quality)
For every read
Reported by the aligners
Consensus quality (variant call quality)
For every genomic locus
Reported by the variant callers
Quality scores
Phred scores
Published in 1998
Initially developed for human genome project
Widely used to characterize the quality of DNA sequence
Q= -10log10§
Q = 10; P = 0.1; acc = 90%
Q = 20; P = 0.01; acc = 99%
…
Sequence alignment
A way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity
Helps in inferring functional, structural, or evolutionary relationships between the sequences
Goal: find out the best matching sequences
Global vs Local

Alignment theories
Scoring matrix, or penalty scheme
Protein: PAM and BLOSUM
DNA/RNA
Match = 1
Mismatch = 0
Gap
d = 3 (gap opening)
e = 0.1 (gap extension)

Global alignment
must account for all characters of each sequence
Needleman-Wunsch algorithm
Local alignment
accounts for only a continuous portion of each sequence
Smith-Waterman algorithm
Searching can start/end anywhere
Fast alignment for short reads
Short reads aligner
Major challenge: Going through 1 trillion times (reads) dynamic programming is not practical
Strategy: making a dictionary (index)
Problem: Making a 50-nt index is too huge: 450 = 1.3*1030
Things to consider
Features: Short and massive amounts
Cost
Speed, Resources required (memory)
Alignment quality
Gaps allowed?
Information considered
Base sequence quality considered?
Accuracy
Short reads aligner strategies
Three common strategies
Hash table
Seed-extend paradigm

Space allowance
Suffix/Prefix tree
Suffix array
Burrows-Wheeler transformation
==Merge sorting ==(not commonly used)
Hash table - based algorithm
Algorithms
Hashing reads
Eland, MAQ, Mosaik…
Hashing reference genome
BFAST, Mosaik, SOAP
Hash table - space allowance
Perfect match is straightforward, but not useful to identify genetic variants
Solution: using multiple indices that allow mismatches

More than one way to build mask
Allow 1-nt mismatch m: seed length;
w: weight (number of counted nt)
k: number of allowed mismatches
n: number of indices

What’s the best mask design?
The seed weight w too small—— too many false positives that slow down the mapping process
The seed weight w too higher—— more seeds needed to achieve full sensitivity —— more memory
Optimal mask design: Lin et al. Bioinformatics (2008): ZOOM! Zillions of oligos mapped
mismatch k = 2

Suffix/prefix tree
Problems of hash table based strategy:
Alignment to multiple identical copies of a substring in the reference mast be performed for each copy.
当基因组中重复序列过多时,使得align的速度变慢
Suffix/prefix tree (trie) can handle this well
fast query, O(n), where n is the length of the query sequence.
Nodes: suffix array interval of …
Substring form node to root
Burrows-Wheeler Transformation (BWA, bowtie use)
Developed in 1994
It is an algorithm developed for data compression, such as bzip2

All the letters remain the same value
Order changed
If the original string had several substrings that occurred often, the transformed string will have several places where a single character is repeated multiple times in a row
Easier for compression

Burrows M, Wheeler DJ A block sorting lossless data compression algorithm. 1994
https://baike.baidu.com/item/Burrows-Wheeler%E5%8F%98%E6%8D%A2/752836?fr=aladdin
Two questions
Why is it reversible?
T = acaacg$
BWT(T) = gc$aaac
In any language that has a sort() function, usually, you cannot find an unsort()
How can this strange transformation help me for sequence alignment?
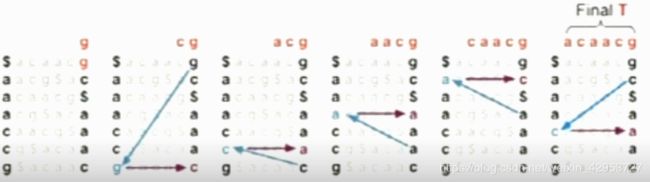
Query using BWT (FM index)
To match Q in T using BWT(T), repeatedly apply rule:
top = LF(top, qc); bot = LF(bot, qc)
Where qc is the next character in Q(right-to-left) and LF(i,qc) maps row i to the row whose first character corresponds to i’s last character as if it were qc
Checkpointing in FM Index
Solution: per-calculate cumulative counts for A/C/G/T up to periodic checkpoints in BWT
Rows to Reference Positions
Bowtie marks every 32nd row by default(configurable)
FM Index is Small
Entire FM Index on DNA reference consists of:
BWT(same size as T)
Checkpoints(~15% size of T)
SA sample(~50% size of T)
Total: ~1.65x the size of T
Alignment tools that uses BWT
BOWTIE
Langmead et al., Genome Biology, 2009
Super efficient, super fast
Biggest problem: Do not allow gaps
Used by many other popular downstream tools, like TOPHAT
BWA
Li H and Durbin R. (2009) Bioinformatics
Allow gaps
Two versions
BWA-short: shorter than 200bp
BWA-SW: longer sequences, up to 100kb
Sequencing error is considered
Evaluations for alignment
Straight Smith-Waterman scores
BFAST
A phred-scale mapping quality calculation
Used in MAQ
Modified in BOWTIE and BWA
A posterior probability p
z: read
x: reference sequence
u: mismatch position
Base quality score of
A: 20 (p=0.01) B: 10 (p=0.1)
The probability that this read is “perfect match” is both A and B are not sequencing error
p=0.01*0.1=0.001
Every other read will have very low p
So, if mapping is unique, p ~= 1
Q(u|x,z) = -10log10[1-p(u|x,z)]
如果mismatch多于2个,输出结果不好
Accuracy:
Sensitivity(recall);
Specificify
Selectivity
Other factors may be included in the evaluation, but sometime should be evaluated independently.
Number of mismatches allowed
Number of gaps allowed
Unique mapping
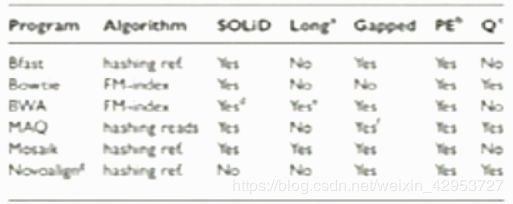
Comparison among short read aligners
David M. et al. SHRiMP2, Bioinformatics, 2001
Most commonly used aligners: BFAST, BWA, Bowtie vs. SHRiMP2

File format
Files
FASTQ
Raw sequences
Format variations:
.qseq (Illumina)
.csfasta + .qual (SOLiD)
SAM/BAM
Sequence alignment
.export —— all alignments, not sorted, including not aligned reads (Illumina)
.sorted —— unique aligned reads (Illumina)
VCF
Variants
.pg —— personal genome SNP (UCSC GB)


SAM/BAM
Li et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009
Reference document: http://samtools.sourceforge.net/SAM1.pdf
BAM is a binary (indexable, more compact) representation of SAM
Two components
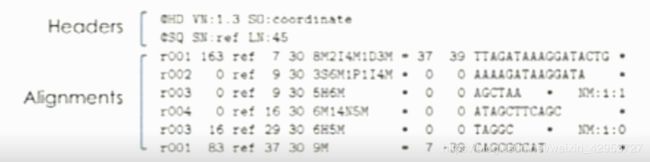
The headers
@HD: File information
format version, sorting order of the alignment
@SQ: Reference sequence information
Reference name, length, assembly identifier, species,…
@RG: Reads group information
RG ID, sequencing center, date, flow order, library, …
@PG: Program
Name and command for alignment program,…
@CO: Additional text comments

1-base coordinate system
SAM, GFF, Wiggle files
First base of a seq is 1
A region is a closed interval
Region between 3nd and 7th nucleotide: [3,7]
0-base coordinate system
BAM, BED, PSL files
First base of a seq is 0
A region is a half closed half open interval
Region between 3nd and 7th nucleotide: [2,7]
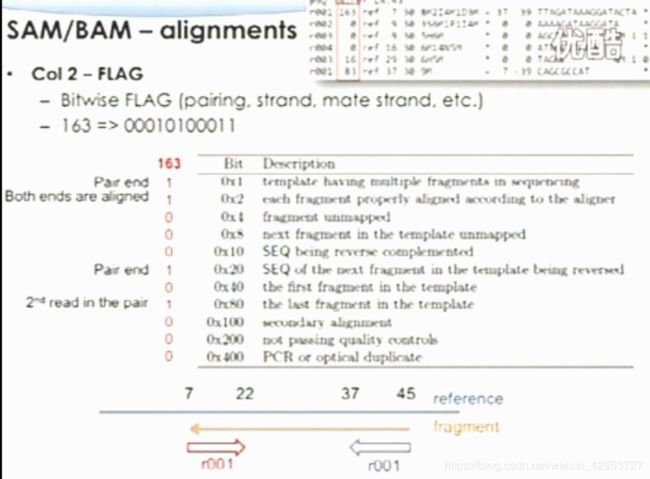
Col 6—— CIGAR operators
M: match/mismatch
I: insertion
D: deletion (8M2I4M1D3M)
H: hard-clipped( unaligned, can only be first and/or last operation)
S: soft-clipped (can have H operations between them and the ends of the CIGAR string)
P: padding
N: skip
Len = M + I + S(H)
Col 9—— inferred fragment size(signed observed template length)
Only for pair-end ( 0 for single-end)
Number of bases from the leftmost mapped base to the rightmost mapped base
Leftmost read(+), rightmost read(-)
SAM/BAM——optional fields
Format
TAG:TYPE:VALUE(6 types: A,i, f, Z, H, B)
Example: NM:I:2
35 pre-defined fields in the v 1.4-r962 version
Allow users to define
Selected fields


BAM( and BAI-indexing)
Binary format of SAM, compressed in the BGZF format
Can be indexed to achieve fast retrieval of alignments overlapping a specified region without going through the whole alignments
must be sorted by the reference ID and then the leftmost coordinate before indexing.
Updated information: http://samtools.sourceforge.net/SAM1.pdf

